Gene
KWMTBOMO14155
Pre Gene Modal
BGIBMGA011409
Annotation
PREDICTED:_scavenger_receptor_class_B_member_1-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.214
Sequence
CDS
ATGTTTTTTGTACAATCGGCATTCACAATACTGGCCAGTTCATATGGAAGCGAATTATTTGTAAAGAAGACTGTTCACCAATACCTTTGGGATTTTAGAGAACCAGTTTTGGATTTATCCAAGAACTTAGCTCCGGGTCTGGTGCCCGTCAACAATATGGGCATATTGGCTAGGATATACGCCAATTTCCAAGACGAGATGACGGTGAAAATAGGTCCGCGCTGGGGCCACGACGAGTTCTTTCAAATAGACAGGTTCTGGGGGAACCCGCAGCTGCCTGGATACGATCCCGAGGTGTGTCCAGATCGAATCGCGGGTTCTACTGAAGGCGTAATGTACCGGCAGCGCCTCACAAAAAACGATACCTTATTATACTGGAGGAAGACTGTCTGCAAAATTATGCCACTTTACTTTGATAGCGAAATGACCTTAGAAGGAGTGCCGGTGTACAGATACAACTTGTCGGAGAGTGTGCACGAACGTCTGACGAACGGAACAGACTGTTACGCCAGCAGCCCGACCTTGCCGGACGGCGTCAGTGACGCTTCGAAATGCTATTACAATTTCCCAATGGTGTCGTCCTATCCACATTTCTACGCGGGCGGTGTTCTCAAAGACCAGTTTGTGACCGGCCTCAAGCCTGATAGAATTAAGCACAATTCTTACGTGATTGTTGAACCGCTAACGGGTACTCCGTTTCACTCTGTGGCGCGAATGCAAAGTAATCTGCACATTAGAGATTTGTCAGGGTTCAATTACAACTATGACAAGCTCTCGAATTTAATATTACCACTTTTCTGGGCAGAATATAATCAAGAAAATCTGCCTTCGAAAATAAAAAACGTGATATACTTCATGGTCGTGATACTGCCGCCCCTAACGATCATAGTTTTATCAATAATAATAATATTAGGTTTCTATTTAATAGCGAAAAACGTATACGAAACGAAACTGCGTTACGACAAAATAAAAACGTTATTGCATTTCAATAGTGACGCTTCGAACGTGAAGACGAATACAATATTTAATTATGAGAAAGAAACGTTCTTGAAGAGTCCTAGTTAA
Protein
MFFVQSAFTILASSYGSELFVKKTVHQYLWDFREPVLDLSKNLAPGLVPVNNMGILARIYANFQDEMTVKIGPRWGHDEFFQIDRFWGNPQLPGYDPEVCPDRIAGSTEGVMYRQRLTKNDTLLYWRKTVCKIMPLYFDSEMTLEGVPVYRYNLSESVHERLTNGTDCYASSPTLPDGVSDASKCYYNFPMVSSYPHFYAGGVLKDQFVTGLKPDRIKHNSYVIVEPLTGTPFHSVARMQSNLHIRDLSGFNYNYDKLSNLILPLFWAEYNQENLPSKIKNVIYFMVVILPPLTIIVLSIIIILGFYLIAKNVYETKLRYDKIKTLLHFNSDASNVKTNTIFNYEKETFLKSPS
Summary
Similarity
Belongs to the CD36 family.
Uniprot
A0A3S2TT24
A0A2W1BBX7
A0A2A4JYP1
A0A2H1V6S5
A0A0N1IQ22
A0A194PUK3
+ More
A0A1W4WNH6 D2A5M8 A0A1B6GRE8 A0A2S2PPX0 A0A2H8TL11 A0A1B6MHQ7 J9K8T5 N6T4A9 J3JYX0 A0A224XAE1 A0A1S4EVC1 Q17PS3 A0A1J1ITJ7 A0A1B0DID4 A0A182TFD4 A0A182WS41 A0A182LHW7 Q7PPV3 A0A182HXT7 A0A1S4GMD3 A0A182JQ14 A0A146KWQ3 A0A182JEV2 E0W152 A0A336MXV2 A0A336MGZ2 A0A084VUD7 A0A0A9Z9V8 A0A182QYU6 A0A182NV31 A0A2M4CLW3 A0A2M4BN34 A0A2M4BMX8 W5JL92 A0A2J7PW74 A0A2M4AQ52 A0A182WMJ9 A0A2M4AQ97 A0A182FCS4 A0A1B0CMS0 A0A067RLQ9 A0A182RQG0 A0A2S2QPK3 J3JXK3 N6TD95 A0A182VK38 A0A2J7PNU5 A0A067QYX6 A0A1B6CDS3 A0A1B6E1D3 T1HPT0 A0A2S2QTV0 A0A3Q9EJV0 A0A2J7PW76 N6TZF9 N6T293 A0A023F2D4 A0A2S2Q087 A0A0K8WGU0 A0A034VI50 A0A3Q9EL58 A0A026X1F9 A0A0A1XC80 A0A0N0U3K8 B4JU08 A0A1W4WGT2 E0W134 A0A3S2NFK2 A0A2S2QNL3 A0A067RCA5 A0A151I684 A0A139WG73 A0A0M3QVG3 A0A2H1WLC8 A0A212EY21 Q7Q7R9 Q17HA4 B4KQT0 Q9W0X1 B4IHC0
A0A1W4WNH6 D2A5M8 A0A1B6GRE8 A0A2S2PPX0 A0A2H8TL11 A0A1B6MHQ7 J9K8T5 N6T4A9 J3JYX0 A0A224XAE1 A0A1S4EVC1 Q17PS3 A0A1J1ITJ7 A0A1B0DID4 A0A182TFD4 A0A182WS41 A0A182LHW7 Q7PPV3 A0A182HXT7 A0A1S4GMD3 A0A182JQ14 A0A146KWQ3 A0A182JEV2 E0W152 A0A336MXV2 A0A336MGZ2 A0A084VUD7 A0A0A9Z9V8 A0A182QYU6 A0A182NV31 A0A2M4CLW3 A0A2M4BN34 A0A2M4BMX8 W5JL92 A0A2J7PW74 A0A2M4AQ52 A0A182WMJ9 A0A2M4AQ97 A0A182FCS4 A0A1B0CMS0 A0A067RLQ9 A0A182RQG0 A0A2S2QPK3 J3JXK3 N6TD95 A0A182VK38 A0A2J7PNU5 A0A067QYX6 A0A1B6CDS3 A0A1B6E1D3 T1HPT0 A0A2S2QTV0 A0A3Q9EJV0 A0A2J7PW76 N6TZF9 N6T293 A0A023F2D4 A0A2S2Q087 A0A0K8WGU0 A0A034VI50 A0A3Q9EL58 A0A026X1F9 A0A0A1XC80 A0A0N0U3K8 B4JU08 A0A1W4WGT2 E0W134 A0A3S2NFK2 A0A2S2QNL3 A0A067RCA5 A0A151I684 A0A139WG73 A0A0M3QVG3 A0A2H1WLC8 A0A212EY21 Q7Q7R9 Q17HA4 B4KQT0 Q9W0X1 B4IHC0
Pubmed
28756777
26354079
18362917
19820115
23537049
22516182
+ More
17510324 20966253 12364791 26823975 20566863 24438588 25401762 20920257 23761445 24845553 30542294 25474469 25348373 24508170 30249741 25830018 17994087 22118469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
17510324 20966253 12364791 26823975 20566863 24438588 25401762 20920257 23761445 24845553 30542294 25474469 25348373 24508170 30249741 25830018 17994087 22118469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
RSAL01000005
RVE54417.1
KZ150337
PZC71324.1
NWSH01000384
PCG76798.1
+ More
ODYU01000781 SOQ36092.1 KQ460036 KPJ18404.1 KQ459593 KPI96429.1 KQ971345 EFA05690.2 GECZ01004780 JAS64989.1 GGMR01018902 MBY31521.1 GFXV01002173 MBW13978.1 GEBQ01004489 JAT35488.1 ABLF02032277 APGK01052766 APGK01052767 APGK01052768 KB741216 ENN72458.1 BT128451 KB632404 AEE63408.1 ERL95040.1 GFTR01007079 JAW09347.1 CH477189 EAT48768.1 CVRI01000059 CRL03535.1 AJVK01015508 AAAB01008905 EAA09702.6 APCN01005347 APCN01005348 GDHC01019419 JAP99209.1 DS235866 EEB19358.1 UFQT01001469 SSX30708.1 UFQT01001251 SSX29652.1 ATLV01016722 KE525103 KFB41581.1 GBHO01003491 JAG40113.1 AXCN02001921 GGFL01002091 MBW66269.1 GGFJ01005232 MBW54373.1 GGFJ01005231 MBW54372.1 ADMH02000839 ETN64886.1 NEVH01020937 PNF20587.1 GGFK01009586 MBW42907.1 GGFK01009571 MBW42892.1 AJWK01019130 AJWK01019131 AJWK01019132 AJWK01019133 AJWK01019134 KK852602 KDR20475.1 GGMS01010454 MBY79657.1 BT127972 AEE62934.1 APGK01042408 APGK01042409 APGK01042410 KB741005 ENN75688.1 NEVH01023284 PNF18020.1 KK853094 KDR11500.1 GEDC01025692 JAS11606.1 GEDC01005560 JAS31738.1 ACPB03008273 GGMS01011955 MBY81158.1 MK049031 AZQ24972.1 PNF20584.1 APGK01055245 KB741261 KB632413 ENN71632.1 ERL95261.1 ENN71633.1 ERL95262.1 GBBI01003543 JAC15169.1 GGMS01001943 MBY71146.1 GDHF01001943 JAI50371.1 GAKP01016788 JAC42164.1 MK049029 AZQ24970.1 KK107031 QOIP01000008 EZA62097.1 RLU19322.1 GBXI01005740 JAD08552.1 KQ435878 KOX69994.1 CH916374 EDV91587.1 EEB19340.1 RSAL01000135 RVE46258.1 GGMS01010060 MBY79263.1 KDR20474.1 KQ978495 KYM93627.1 KQ971348 KYB26889.1 CP012524 ALC42369.1 ODYU01009439 SOQ53869.1 AGBW02011640 OWR46357.1 AAAB01008952 EAA10557.4 CH477251 EAT46038.2 CH933808 EDW09279.1 AE013599 AY069143 AAF47308.1 AAL39288.1 CH480838 EDW49296.1
ODYU01000781 SOQ36092.1 KQ460036 KPJ18404.1 KQ459593 KPI96429.1 KQ971345 EFA05690.2 GECZ01004780 JAS64989.1 GGMR01018902 MBY31521.1 GFXV01002173 MBW13978.1 GEBQ01004489 JAT35488.1 ABLF02032277 APGK01052766 APGK01052767 APGK01052768 KB741216 ENN72458.1 BT128451 KB632404 AEE63408.1 ERL95040.1 GFTR01007079 JAW09347.1 CH477189 EAT48768.1 CVRI01000059 CRL03535.1 AJVK01015508 AAAB01008905 EAA09702.6 APCN01005347 APCN01005348 GDHC01019419 JAP99209.1 DS235866 EEB19358.1 UFQT01001469 SSX30708.1 UFQT01001251 SSX29652.1 ATLV01016722 KE525103 KFB41581.1 GBHO01003491 JAG40113.1 AXCN02001921 GGFL01002091 MBW66269.1 GGFJ01005232 MBW54373.1 GGFJ01005231 MBW54372.1 ADMH02000839 ETN64886.1 NEVH01020937 PNF20587.1 GGFK01009586 MBW42907.1 GGFK01009571 MBW42892.1 AJWK01019130 AJWK01019131 AJWK01019132 AJWK01019133 AJWK01019134 KK852602 KDR20475.1 GGMS01010454 MBY79657.1 BT127972 AEE62934.1 APGK01042408 APGK01042409 APGK01042410 KB741005 ENN75688.1 NEVH01023284 PNF18020.1 KK853094 KDR11500.1 GEDC01025692 JAS11606.1 GEDC01005560 JAS31738.1 ACPB03008273 GGMS01011955 MBY81158.1 MK049031 AZQ24972.1 PNF20584.1 APGK01055245 KB741261 KB632413 ENN71632.1 ERL95261.1 ENN71633.1 ERL95262.1 GBBI01003543 JAC15169.1 GGMS01001943 MBY71146.1 GDHF01001943 JAI50371.1 GAKP01016788 JAC42164.1 MK049029 AZQ24970.1 KK107031 QOIP01000008 EZA62097.1 RLU19322.1 GBXI01005740 JAD08552.1 KQ435878 KOX69994.1 CH916374 EDV91587.1 EEB19340.1 RSAL01000135 RVE46258.1 GGMS01010060 MBY79263.1 KDR20474.1 KQ978495 KYM93627.1 KQ971348 KYB26889.1 CP012524 ALC42369.1 ODYU01009439 SOQ53869.1 AGBW02011640 OWR46357.1 AAAB01008952 EAA10557.4 CH477251 EAT46038.2 CH933808 EDW09279.1 AE013599 AY069143 AAF47308.1 AAL39288.1 CH480838 EDW49296.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000192223
UP000007266
+ More
UP000007819 UP000019118 UP000030742 UP000008820 UP000183832 UP000092462 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075881 UP000075880 UP000009046 UP000030765 UP000075886 UP000075884 UP000000673 UP000235965 UP000075920 UP000069272 UP000092461 UP000027135 UP000075900 UP000075903 UP000015103 UP000053097 UP000279307 UP000053105 UP000001070 UP000078542 UP000092553 UP000007151 UP000009192 UP000000803 UP000001292
UP000007819 UP000019118 UP000030742 UP000008820 UP000183832 UP000092462 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075881 UP000075880 UP000009046 UP000030765 UP000075886 UP000075884 UP000000673 UP000235965 UP000075920 UP000069272 UP000092461 UP000027135 UP000075900 UP000075903 UP000015103 UP000053097 UP000279307 UP000053105 UP000001070 UP000078542 UP000092553 UP000007151 UP000009192 UP000000803 UP000001292
PRIDE
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
A0A3S2TT24
A0A2W1BBX7
A0A2A4JYP1
A0A2H1V6S5
A0A0N1IQ22
A0A194PUK3
+ More
A0A1W4WNH6 D2A5M8 A0A1B6GRE8 A0A2S2PPX0 A0A2H8TL11 A0A1B6MHQ7 J9K8T5 N6T4A9 J3JYX0 A0A224XAE1 A0A1S4EVC1 Q17PS3 A0A1J1ITJ7 A0A1B0DID4 A0A182TFD4 A0A182WS41 A0A182LHW7 Q7PPV3 A0A182HXT7 A0A1S4GMD3 A0A182JQ14 A0A146KWQ3 A0A182JEV2 E0W152 A0A336MXV2 A0A336MGZ2 A0A084VUD7 A0A0A9Z9V8 A0A182QYU6 A0A182NV31 A0A2M4CLW3 A0A2M4BN34 A0A2M4BMX8 W5JL92 A0A2J7PW74 A0A2M4AQ52 A0A182WMJ9 A0A2M4AQ97 A0A182FCS4 A0A1B0CMS0 A0A067RLQ9 A0A182RQG0 A0A2S2QPK3 J3JXK3 N6TD95 A0A182VK38 A0A2J7PNU5 A0A067QYX6 A0A1B6CDS3 A0A1B6E1D3 T1HPT0 A0A2S2QTV0 A0A3Q9EJV0 A0A2J7PW76 N6TZF9 N6T293 A0A023F2D4 A0A2S2Q087 A0A0K8WGU0 A0A034VI50 A0A3Q9EL58 A0A026X1F9 A0A0A1XC80 A0A0N0U3K8 B4JU08 A0A1W4WGT2 E0W134 A0A3S2NFK2 A0A2S2QNL3 A0A067RCA5 A0A151I684 A0A139WG73 A0A0M3QVG3 A0A2H1WLC8 A0A212EY21 Q7Q7R9 Q17HA4 B4KQT0 Q9W0X1 B4IHC0
A0A1W4WNH6 D2A5M8 A0A1B6GRE8 A0A2S2PPX0 A0A2H8TL11 A0A1B6MHQ7 J9K8T5 N6T4A9 J3JYX0 A0A224XAE1 A0A1S4EVC1 Q17PS3 A0A1J1ITJ7 A0A1B0DID4 A0A182TFD4 A0A182WS41 A0A182LHW7 Q7PPV3 A0A182HXT7 A0A1S4GMD3 A0A182JQ14 A0A146KWQ3 A0A182JEV2 E0W152 A0A336MXV2 A0A336MGZ2 A0A084VUD7 A0A0A9Z9V8 A0A182QYU6 A0A182NV31 A0A2M4CLW3 A0A2M4BN34 A0A2M4BMX8 W5JL92 A0A2J7PW74 A0A2M4AQ52 A0A182WMJ9 A0A2M4AQ97 A0A182FCS4 A0A1B0CMS0 A0A067RLQ9 A0A182RQG0 A0A2S2QPK3 J3JXK3 N6TD95 A0A182VK38 A0A2J7PNU5 A0A067QYX6 A0A1B6CDS3 A0A1B6E1D3 T1HPT0 A0A2S2QTV0 A0A3Q9EJV0 A0A2J7PW76 N6TZF9 N6T293 A0A023F2D4 A0A2S2Q087 A0A0K8WGU0 A0A034VI50 A0A3Q9EL58 A0A026X1F9 A0A0A1XC80 A0A0N0U3K8 B4JU08 A0A1W4WGT2 E0W134 A0A3S2NFK2 A0A2S2QNL3 A0A067RCA5 A0A151I684 A0A139WG73 A0A0M3QVG3 A0A2H1WLC8 A0A212EY21 Q7Q7R9 Q17HA4 B4KQT0 Q9W0X1 B4IHC0
PDB
5LGD
E-value=6.8894e-18,
Score=222
Ontologies
GO
PANTHER
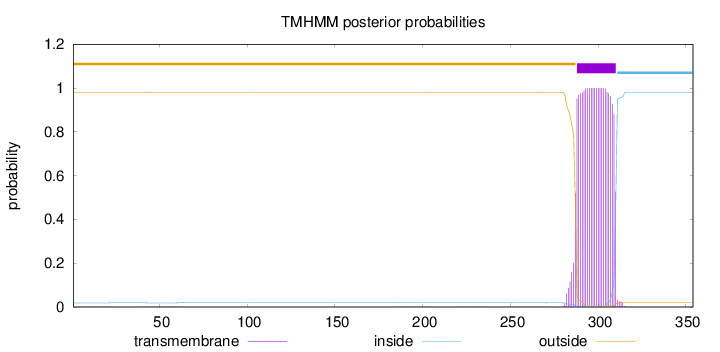
Topology
Length:
354
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.44818
Exp number, first 60 AAs:
0.04658
Total prob of N-in:
0.01904
outside
1 - 287
TMhelix
288 - 310
inside
311 - 354
Population Genetic Test Statistics
Pi
21.071456
Theta
20.313595
Tajima's D
0.180072
CLR
1.699111
CSRT
0.421728913554322
Interpretation
Uncertain
