Gene
KWMTBOMO14147
Pre Gene Modal
BGIBMGA011404
Annotation
Neurofibromin_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.041
Sequence
CDS
ATGGGAACGCAGAAACCTGGAGAATGGGCTAATTCGTTGATAGCGCGTTTCGAAGAACAGCTCCCTTATAGGACGGGAGCACAGAACCTGCACTCAAGGATTAATGAGGAACAATGTAAAGCATGCCTCGTCCAGATAAGCAGATACCGGTTCTCATTAGTCATTTCAGGGCTGACGAAAATACTGCAGAGAGTAAATGAACTGTATCAGCCAACATTCACAAGCAGCATCAATAGACCTCAAACAGAGCTGGAGAAGGGCTACCACGATAGTCTTGTGATAGTCCTGGACACACTTGAGATATGCCTCAGCTCACAGCCCAAGGATACTGCCAAATATGATGAAGCAATGAATGTAAAAATTCTGCTTCGGGAAGTCTGTCAGTTTATGGATTCACCCTAA
Protein
MGTQKPGEWANSLIARFEEQLPYRTGAQNLHSRINEEQCKACLVQISRYRFSLVISGLTKILQRVNELYQPTFTSSINRPQTELEKGYHDSLVIVLDTLEICLSSQPKDTAKYDEAMNVKILLREVCQFMDSP
Summary
Uniprot
A0A194PTX2
H9JPF0
A0A2A4J4I6
A0A0N1IBQ7
A0A2W1BS17
A0A1B6MMK6
+ More
A0A1B6CFS5 A0A2J7Q2H1 A0A2J7PY13 A0A226E5T9 A0A087ULX9 A0A1B6CM43 K7J294 A0A232EX77 A0A023EXJ2 A0A1B6H8S9 A0A1S4FJK1 F4WPG4 A0A0P5N844 E9H0M4 A0A158NR60 A0A182NSE1 A0A182LY02 A0A182XE90 A0A182JYC4 A0A182TUB1 A0A182RYC1 A0A182PSW3 A0A182LC78 A0A182VES4 A0A182W2S9 A0A182HJ32 A0A182Q4E2 E2ADQ4 A0A1B6I0Q1 A0A3L8DWI0 A0A1Y1KHF5 A0A0P6IU49 F5HL00 Q7QBJ9 A0A1Y1KPV6 A0A084WSZ8 Q16Z01 A0A1Y1KHG0 B0WYP5 A0A1Y1KHE9 A0A088A4V7 A0A2A3E8Q2 A0A2M4CMF2 A0A182FL69 A0A2M4A6W8 A0A182XVJ7 A0A139WPT3 A0A139WPE8 A0A1B0D165 A0A3B0KLU3 A0A1B0C857 A0A147BNF4 V5GMC6 A0A2M4A6Z0 W5JE11 A0A0Q9X895 A0A0Q9XA70 A0A0Q9XCE0 A0A0Q9X9D7 B4K6B9 B3LYI6 A0A1W4V911 A0A1W4VKR6 A0A1W4VM54 A0A1J1ITN5 A0A1J1IWR0 A0A1Y3BFY2 A0A0K8VSX4 B5DX59 A0A2P2IEF1 B4GEH6 A0A3B0KSB7 Q1WWH0 A0A1I8PHZ4 A0A1I8PI47 A0A1I8PHU0 A0A3S3PH18 A0A1W4WNP1 B4QW05 B4LYR3 A0A0B4KHX4 A0A0B4KI28 O01397 Q9VBJ2 B3P6C2 B4PSY7 O01398 O01399 Q59DT9 Q8IMS2 A0A1I8N339 B4JGH8 A0A0C9RNP7
A0A1B6CFS5 A0A2J7Q2H1 A0A2J7PY13 A0A226E5T9 A0A087ULX9 A0A1B6CM43 K7J294 A0A232EX77 A0A023EXJ2 A0A1B6H8S9 A0A1S4FJK1 F4WPG4 A0A0P5N844 E9H0M4 A0A158NR60 A0A182NSE1 A0A182LY02 A0A182XE90 A0A182JYC4 A0A182TUB1 A0A182RYC1 A0A182PSW3 A0A182LC78 A0A182VES4 A0A182W2S9 A0A182HJ32 A0A182Q4E2 E2ADQ4 A0A1B6I0Q1 A0A3L8DWI0 A0A1Y1KHF5 A0A0P6IU49 F5HL00 Q7QBJ9 A0A1Y1KPV6 A0A084WSZ8 Q16Z01 A0A1Y1KHG0 B0WYP5 A0A1Y1KHE9 A0A088A4V7 A0A2A3E8Q2 A0A2M4CMF2 A0A182FL69 A0A2M4A6W8 A0A182XVJ7 A0A139WPT3 A0A139WPE8 A0A1B0D165 A0A3B0KLU3 A0A1B0C857 A0A147BNF4 V5GMC6 A0A2M4A6Z0 W5JE11 A0A0Q9X895 A0A0Q9XA70 A0A0Q9XCE0 A0A0Q9X9D7 B4K6B9 B3LYI6 A0A1W4V911 A0A1W4VKR6 A0A1W4VM54 A0A1J1ITN5 A0A1J1IWR0 A0A1Y3BFY2 A0A0K8VSX4 B5DX59 A0A2P2IEF1 B4GEH6 A0A3B0KSB7 Q1WWH0 A0A1I8PHZ4 A0A1I8PI47 A0A1I8PHU0 A0A3S3PH18 A0A1W4WNP1 B4QW05 B4LYR3 A0A0B4KHX4 A0A0B4KI28 O01397 Q9VBJ2 B3P6C2 B4PSY7 O01398 O01399 Q59DT9 Q8IMS2 A0A1I8N339 B4JGH8 A0A0C9RNP7
Pubmed
26354079
19121390
28756777
20075255
28648823
25474469
+ More
17510324 21719571 21292972 21347285 20966253 20798317 30249741 28004739 12364791 24438588 25244985 18362917 19820115 29652888 20920257 23761445 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9115203 17550304 25315136
17510324 21719571 21292972 21347285 20966253 20798317 30249741 28004739 12364791 24438588 25244985 18362917 19820115 29652888 20920257 23761445 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9115203 17550304 25315136
EMBL
KQ459593
KPI96423.1
BABH01009999
BABH01010000
NWSH01003377
PCG66424.1
+ More
KQ460036 KPJ18410.1 KZ149959 PZC76415.1 GEBQ01002878 JAT37099.1 GEDC01024951 JAS12347.1 NEVH01019370 PNF22780.1 NEVH01020852 PNF21215.1 LNIX01000006 OXA52668.1 KK120488 KFM78368.1 GEDC01022860 JAS14438.1 AAZX01001574 AAZX01008897 AAZX01014691 NNAY01001785 OXU22929.1 GBBI01004800 JAC13912.1 GECU01036588 JAS71118.1 GL888243 EGI64059.1 GDIQ01145802 JAL05924.1 GL732581 EFX74727.1 ADTU01023728 ADTU01023729 ADTU01023730 AXCM01000247 APCN01002121 AXCN02000238 GL438820 EFN68416.1 GECU01027208 JAS80498.1 QOIP01000003 RLU24672.1 GEZM01083962 GEZM01083961 GEZM01083957 GEZM01083956 JAV60893.1 GDIQ01001376 JAN93361.1 AAAB01008879 EGK96961.1 EAA08440.4 GEZM01083960 GEZM01083959 JAV60897.1 ATLV01026776 ATLV01026777 ATLV01026778 ATLV01026779 ATLV01026780 KE525419 KFB53342.1 CH477505 EAT39867.1 GEZM01083955 JAV60909.1 DS232196 EDS37155.1 GEZM01083958 JAV60902.1 KZ288323 PBC28105.1 GGFL01002342 MBW66520.1 GGFK01003222 MBW36543.1 KQ971307 KYB29815.1 KYB29814.1 AJVK01021962 AJVK01021963 AJVK01021964 OUUW01000023 SPP89590.1 AJWK01000199 AJWK01000200 AJWK01000201 AJWK01000202 AJWK01000203 GEGO01003091 JAR92313.1 GALX01005699 JAB62767.1 GGFK01003233 MBW36554.1 ADMH02001497 ETN62306.1 CH933806 KRG01444.1 KRG01441.1 KRG01443.1 KRG01442.1 EDW15192.1 CH902617 EDV42901.2 CVRI01000059 CRL03597.1 CRL03598.1 MUJZ01026521 OTF78733.1 GDHF01010327 JAI41987.1 CM000070 EDY68218.3 IACF01006808 LAB72391.1 CH479182 EDW34011.1 SPP89589.1 BT024936 ABE01166.1 NCKU01002548 NCKU01002470 RWS09353.1 RWS09518.1 CM000364 EDX14504.1 CH940650 EDW66990.2 AE014297 AGB96359.1 AGB96358.1 L26500 AAB58977.1 AAF56543.3 CH954182 EDV53592.1 CM000160 EDW98674.2 L26501 AAB58975.1 L26502 AAB58976.1 AAX52994.1 AAN14067.2 CH916369 EDV93675.1 GBYB01010045 JAG79812.1
KQ460036 KPJ18410.1 KZ149959 PZC76415.1 GEBQ01002878 JAT37099.1 GEDC01024951 JAS12347.1 NEVH01019370 PNF22780.1 NEVH01020852 PNF21215.1 LNIX01000006 OXA52668.1 KK120488 KFM78368.1 GEDC01022860 JAS14438.1 AAZX01001574 AAZX01008897 AAZX01014691 NNAY01001785 OXU22929.1 GBBI01004800 JAC13912.1 GECU01036588 JAS71118.1 GL888243 EGI64059.1 GDIQ01145802 JAL05924.1 GL732581 EFX74727.1 ADTU01023728 ADTU01023729 ADTU01023730 AXCM01000247 APCN01002121 AXCN02000238 GL438820 EFN68416.1 GECU01027208 JAS80498.1 QOIP01000003 RLU24672.1 GEZM01083962 GEZM01083961 GEZM01083957 GEZM01083956 JAV60893.1 GDIQ01001376 JAN93361.1 AAAB01008879 EGK96961.1 EAA08440.4 GEZM01083960 GEZM01083959 JAV60897.1 ATLV01026776 ATLV01026777 ATLV01026778 ATLV01026779 ATLV01026780 KE525419 KFB53342.1 CH477505 EAT39867.1 GEZM01083955 JAV60909.1 DS232196 EDS37155.1 GEZM01083958 JAV60902.1 KZ288323 PBC28105.1 GGFL01002342 MBW66520.1 GGFK01003222 MBW36543.1 KQ971307 KYB29815.1 KYB29814.1 AJVK01021962 AJVK01021963 AJVK01021964 OUUW01000023 SPP89590.1 AJWK01000199 AJWK01000200 AJWK01000201 AJWK01000202 AJWK01000203 GEGO01003091 JAR92313.1 GALX01005699 JAB62767.1 GGFK01003233 MBW36554.1 ADMH02001497 ETN62306.1 CH933806 KRG01444.1 KRG01441.1 KRG01443.1 KRG01442.1 EDW15192.1 CH902617 EDV42901.2 CVRI01000059 CRL03597.1 CRL03598.1 MUJZ01026521 OTF78733.1 GDHF01010327 JAI41987.1 CM000070 EDY68218.3 IACF01006808 LAB72391.1 CH479182 EDW34011.1 SPP89589.1 BT024936 ABE01166.1 NCKU01002548 NCKU01002470 RWS09353.1 RWS09518.1 CM000364 EDX14504.1 CH940650 EDW66990.2 AE014297 AGB96359.1 AGB96358.1 L26500 AAB58977.1 AAF56543.3 CH954182 EDV53592.1 CM000160 EDW98674.2 L26501 AAB58975.1 L26502 AAB58976.1 AAX52994.1 AAN14067.2 CH916369 EDV93675.1 GBYB01010045 JAG79812.1
Proteomes
UP000053268
UP000005204
UP000218220
UP000053240
UP000235965
UP000198287
+ More
UP000054359 UP000002358 UP000215335 UP000007755 UP000000305 UP000005205 UP000075884 UP000075883 UP000076407 UP000075881 UP000075902 UP000075900 UP000075885 UP000075882 UP000075903 UP000075920 UP000075840 UP000075886 UP000000311 UP000279307 UP000007062 UP000030765 UP000008820 UP000002320 UP000005203 UP000242457 UP000069272 UP000076408 UP000007266 UP000092462 UP000268350 UP000092461 UP000000673 UP000009192 UP000007801 UP000192221 UP000183832 UP000001819 UP000008744 UP000095300 UP000285301 UP000192223 UP000000304 UP000008792 UP000000803 UP000008711 UP000002282 UP000095301 UP000001070
UP000054359 UP000002358 UP000215335 UP000007755 UP000000305 UP000005205 UP000075884 UP000075883 UP000076407 UP000075881 UP000075902 UP000075900 UP000075885 UP000075882 UP000075903 UP000075920 UP000075840 UP000075886 UP000000311 UP000279307 UP000007062 UP000030765 UP000008820 UP000002320 UP000005203 UP000242457 UP000069272 UP000076408 UP000007266 UP000092462 UP000268350 UP000092461 UP000000673 UP000009192 UP000007801 UP000192221 UP000183832 UP000001819 UP000008744 UP000095300 UP000285301 UP000192223 UP000000304 UP000008792 UP000000803 UP000008711 UP000002282 UP000095301 UP000001070
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A194PTX2
H9JPF0
A0A2A4J4I6
A0A0N1IBQ7
A0A2W1BS17
A0A1B6MMK6
+ More
A0A1B6CFS5 A0A2J7Q2H1 A0A2J7PY13 A0A226E5T9 A0A087ULX9 A0A1B6CM43 K7J294 A0A232EX77 A0A023EXJ2 A0A1B6H8S9 A0A1S4FJK1 F4WPG4 A0A0P5N844 E9H0M4 A0A158NR60 A0A182NSE1 A0A182LY02 A0A182XE90 A0A182JYC4 A0A182TUB1 A0A182RYC1 A0A182PSW3 A0A182LC78 A0A182VES4 A0A182W2S9 A0A182HJ32 A0A182Q4E2 E2ADQ4 A0A1B6I0Q1 A0A3L8DWI0 A0A1Y1KHF5 A0A0P6IU49 F5HL00 Q7QBJ9 A0A1Y1KPV6 A0A084WSZ8 Q16Z01 A0A1Y1KHG0 B0WYP5 A0A1Y1KHE9 A0A088A4V7 A0A2A3E8Q2 A0A2M4CMF2 A0A182FL69 A0A2M4A6W8 A0A182XVJ7 A0A139WPT3 A0A139WPE8 A0A1B0D165 A0A3B0KLU3 A0A1B0C857 A0A147BNF4 V5GMC6 A0A2M4A6Z0 W5JE11 A0A0Q9X895 A0A0Q9XA70 A0A0Q9XCE0 A0A0Q9X9D7 B4K6B9 B3LYI6 A0A1W4V911 A0A1W4VKR6 A0A1W4VM54 A0A1J1ITN5 A0A1J1IWR0 A0A1Y3BFY2 A0A0K8VSX4 B5DX59 A0A2P2IEF1 B4GEH6 A0A3B0KSB7 Q1WWH0 A0A1I8PHZ4 A0A1I8PI47 A0A1I8PHU0 A0A3S3PH18 A0A1W4WNP1 B4QW05 B4LYR3 A0A0B4KHX4 A0A0B4KI28 O01397 Q9VBJ2 B3P6C2 B4PSY7 O01398 O01399 Q59DT9 Q8IMS2 A0A1I8N339 B4JGH8 A0A0C9RNP7
A0A1B6CFS5 A0A2J7Q2H1 A0A2J7PY13 A0A226E5T9 A0A087ULX9 A0A1B6CM43 K7J294 A0A232EX77 A0A023EXJ2 A0A1B6H8S9 A0A1S4FJK1 F4WPG4 A0A0P5N844 E9H0M4 A0A158NR60 A0A182NSE1 A0A182LY02 A0A182XE90 A0A182JYC4 A0A182TUB1 A0A182RYC1 A0A182PSW3 A0A182LC78 A0A182VES4 A0A182W2S9 A0A182HJ32 A0A182Q4E2 E2ADQ4 A0A1B6I0Q1 A0A3L8DWI0 A0A1Y1KHF5 A0A0P6IU49 F5HL00 Q7QBJ9 A0A1Y1KPV6 A0A084WSZ8 Q16Z01 A0A1Y1KHG0 B0WYP5 A0A1Y1KHE9 A0A088A4V7 A0A2A3E8Q2 A0A2M4CMF2 A0A182FL69 A0A2M4A6W8 A0A182XVJ7 A0A139WPT3 A0A139WPE8 A0A1B0D165 A0A3B0KLU3 A0A1B0C857 A0A147BNF4 V5GMC6 A0A2M4A6Z0 W5JE11 A0A0Q9X895 A0A0Q9XA70 A0A0Q9XCE0 A0A0Q9X9D7 B4K6B9 B3LYI6 A0A1W4V911 A0A1W4VKR6 A0A1W4VM54 A0A1J1ITN5 A0A1J1IWR0 A0A1Y3BFY2 A0A0K8VSX4 B5DX59 A0A2P2IEF1 B4GEH6 A0A3B0KSB7 Q1WWH0 A0A1I8PHZ4 A0A1I8PI47 A0A1I8PHU0 A0A3S3PH18 A0A1W4WNP1 B4QW05 B4LYR3 A0A0B4KHX4 A0A0B4KI28 O01397 Q9VBJ2 B3P6C2 B4PSY7 O01398 O01399 Q59DT9 Q8IMS2 A0A1I8N339 B4JGH8 A0A0C9RNP7
Ontologies
GO
PANTHER
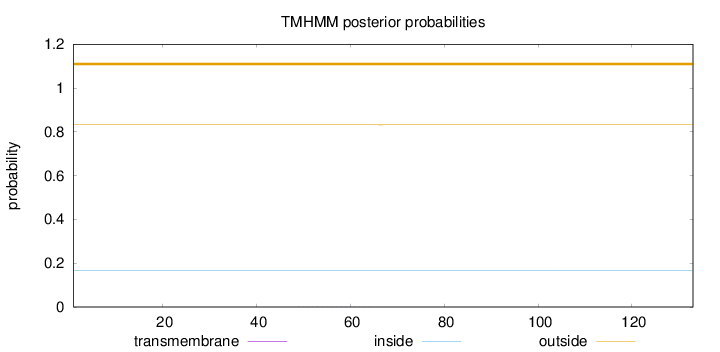
Topology
Length:
133
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00316
Exp number, first 60 AAs:
0.00216
Total prob of N-in:
0.16807
outside
1 - 133
Population Genetic Test Statistics
Pi
17.704591
Theta
19.25099
Tajima's D
-0.506988
CLR
0.635038
CSRT
0.243737813109345
Interpretation
Uncertain
