Gene
KWMTBOMO14146 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011186
Annotation
PREDICTED:_sorting_nexin-6_isoform_X3_[Bombyx_mori]
Full name
Sorting nexin
Location in the cell
Cytoplasmic Reliability : 1.613 Nuclear Reliability : 1.91
Sequence
CDS
ATGATGGACTGTGTGGAAGATTCATCGCAGGACCCTTTGTCAACTGTTCCAGTGGCTGATCCAGCCCCAGCTACCATGGACACAAAAGTCGAGAAAAAGAAGCCAAATGAGAATGTCTCTCTAGCTGATAATAGTCTTCTGGTAGACATATCAGATGCTCTAAGCGAAAAAGAAAAAGTCAAGTTCACTGTTCACACCAAAACAACTTTACCGGAGTTCCAAAAATCAGAGTTCATAGTAGTACGTCAGCATGAAGAGTTTGTCTGGCTACATGACCGTTATGAAGAGAATGAAGAATATGCTGGTTATATAATCCCTCCAGCACCTCCTCGTCCTGATTTCGATGCCTCCCGCGAGAAGCTACAGCGACTGGGCGAGGGGGAAGGAGCCCTCACTAGAGAAGAGTTCCTCAAGATGAAGGAGGAGTTGGAAGACGAGTACCTGGCGACCTTTAAGAAGACGGTGGCGATGCACGAAGTATTCCTCCAGCGCCTGGCTGCTCATCCTGTCTTCCGCAGGGATTCTCATCTCAGGGTATTCCTGGAGTACGAACAGGATCTCTGTGCCAAGCCAAGAGGCAAGATGGACCTCATTGGTGGACTGGTTCGGTCAATGACAACAACCACTGATGAGATATACCTCGGGGCGACCGTGCGAGACGTGAACGACTTCTTCGAGCAGGAGACGTCGTTTCTGCAAGAATACTACTCGCACCTCAAGGAGGCAGTCGCTAAAGTCGACCGTATGACGTCGAAGCACAAAGAAGTGGCGGACGCTCACATCAAGCTGTCGTCTTGCATAACTCAGCTCGCGACCAGAGAGCAGCCTCCCATGGAGAGGTTCCTCACGAAGGCCTCCGAGACATTTGACAAGTGCCGGAAAATCGAGGGTCGCATGGCGTCCGACCAGGACCTGAAGCTGGCGGACACTCTCCGGTACTACATGAGAGACGCTCACGCAGCGAAAGCGGTGCTCGTGAGGAGACTCAGGTGTCTAGCTGCCTACGAAGCAGCCAACAGGAACTTGGAGAAGGCGAGAGCGAAGAATAAAGATGTTCACGCTGCCGAACAAGGTCAGGCGGAGGCGTGCGCTAAATTCGAGCAGCTGTCCGCGAGGGCCAGGGAGGAGCTCATCGACTTCAGGACCAGACGGGTGGCCGCCTTCAGGAAAAGCCTTATAGAACTAGCGGAATTGGAGATAAAGCACGCGAGGTCACAGCAGGAGTTGTTTAGGAAATCACTTCAAGTGCTTAGGGAGTGTCAGTGA
Protein
MMDCVEDSSQDPLSTVPVADPAPATMDTKVEKKKPNENVSLADNSLLVDISDALSEKEKVKFTVHTKTTLPEFQKSEFIVVRQHEEFVWLHDRYEENEEYAGYIIPPAPPRPDFDASREKLQRLGEGEGALTREEFLKMKEELEDEYLATFKKTVAMHEVFLQRLAAHPVFRRDSHLRVFLEYEQDLCAKPRGKMDLIGGLVRSMTTTTDEIYLGATVRDVNDFFEQETSFLQEYYSHLKEAVAKVDRMTSKHKEVADAHIKLSSCITQLATREQPPMERFLTKASETFDKCRKIEGRMASDQDLKLADTLRYYMRDAHAAKAVLVRRLRCLAAYEAANRNLEKARAKNKDVHAAEQGQAEACAKFEQLSARAREELIDFRTRRVAAFRKSLIELAELEIKHARSQQELFRKSLQVLRECQ
Summary
Description
Involved in several stages of intracellular trafficking.
Similarity
Belongs to the sorting nexin family.
Uniprot
H9JNT2
A0A2A4J630
A0A0N1PHK9
A0A2A4J7N3
A0A1E1WK18
A0A212EQY3
+ More
A0A194PSY3 A0A1Q3FA85 A0A1Q3FAK8 A0A1L8E392 A0A182HYN2 A0A1L8E2Z9 A0A182UCR1 A0A182XIJ6 A0A182KZK3 Q7QA12 A0A1L8E2Z0 A0A1Q3FBQ8 A0A182LV83 A0A182W3F2 A0A182NGE7 A0A182JXK9 A0A182UPX8 A0A182JM65 A0A182PP85 A0A182H072 Q0IG11 A0A1S4F5L1 A0A084WA11 A0A182Y8T2 A0A182GPM3 A0A154P9I3 A0A182RZK6 A0A0L7R820 A0A1B0GHQ2 E2BGD2 A0A310SE47 A0A195D7R9 F4W809 A0A195BPH5 A0A336M9Q2 A0A2M4AEJ7 E2AKQ5 A0A195C748 A0A0J7LB69 A0A195EZE3 A0A1B6EY91 E9J529 A0A026W6Y9 A0A182FSS6 A0A023ES73 A0A087ZJV1 A0A1B6HL57 V9IG08 A0A2M4BPK5 A0A2M3Z5W4 A0A0M9AB75 A0A336M9F1 A0A158NDS6 A0A0C9PY51 A0A2J7Q7M5 A0A1J1HUJ8 A0A1B6L960 B0WDK0 A0A0C9R0L5 A0A3L8E079 K7J5J2 A0A151X475 A0A0J9QYG1 B4NWU6 B4HYH0 W8BE42 Q9VLQ9 W5J2Z9 B3N726 A0A0P4VZH5 T1HLS5 D6WA63 A0A1W4V7T4 A0A1Y1MFI5 A0A224XDD8 B4KGX5 B4MVY1 A0A069DYW9 B4LUA9 A0A2A3EIC4 A0A3B0JCK1 A0A2J7Q7M1 A0A088ATA4 A0A0M4ENG3 Q29MM3 A0A232FHJ3 A0A0A9VTD2 A0A067RNM3 A0A0J9QY14 M9NDI3 A0A0V0G5Q3 A0A0R1DKR4 A0A0Q5WBC1 W8B9F4
A0A194PSY3 A0A1Q3FA85 A0A1Q3FAK8 A0A1L8E392 A0A182HYN2 A0A1L8E2Z9 A0A182UCR1 A0A182XIJ6 A0A182KZK3 Q7QA12 A0A1L8E2Z0 A0A1Q3FBQ8 A0A182LV83 A0A182W3F2 A0A182NGE7 A0A182JXK9 A0A182UPX8 A0A182JM65 A0A182PP85 A0A182H072 Q0IG11 A0A1S4F5L1 A0A084WA11 A0A182Y8T2 A0A182GPM3 A0A154P9I3 A0A182RZK6 A0A0L7R820 A0A1B0GHQ2 E2BGD2 A0A310SE47 A0A195D7R9 F4W809 A0A195BPH5 A0A336M9Q2 A0A2M4AEJ7 E2AKQ5 A0A195C748 A0A0J7LB69 A0A195EZE3 A0A1B6EY91 E9J529 A0A026W6Y9 A0A182FSS6 A0A023ES73 A0A087ZJV1 A0A1B6HL57 V9IG08 A0A2M4BPK5 A0A2M3Z5W4 A0A0M9AB75 A0A336M9F1 A0A158NDS6 A0A0C9PY51 A0A2J7Q7M5 A0A1J1HUJ8 A0A1B6L960 B0WDK0 A0A0C9R0L5 A0A3L8E079 K7J5J2 A0A151X475 A0A0J9QYG1 B4NWU6 B4HYH0 W8BE42 Q9VLQ9 W5J2Z9 B3N726 A0A0P4VZH5 T1HLS5 D6WA63 A0A1W4V7T4 A0A1Y1MFI5 A0A224XDD8 B4KGX5 B4MVY1 A0A069DYW9 B4LUA9 A0A2A3EIC4 A0A3B0JCK1 A0A2J7Q7M1 A0A088ATA4 A0A0M4ENG3 Q29MM3 A0A232FHJ3 A0A0A9VTD2 A0A067RNM3 A0A0J9QY14 M9NDI3 A0A0V0G5Q3 A0A0R1DKR4 A0A0Q5WBC1 W8B9F4
Pubmed
19121390
26354079
22118469
20966253
12364791
14747013
+ More
17210077 26483478 17510324 24438588 25244985 20798317 21719571 21282665 24508170 24945155 20920257 21347285 30249741 20075255 22936249 17994087 17550304 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 18362917 19820115 28004739 26334808 15632085 28648823 25401762 26823975 24845553
17210077 26483478 17510324 24438588 25244985 20798317 21719571 21282665 24508170 24945155 20920257 21347285 30249741 20075255 22936249 17994087 17550304 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 18362917 19820115 28004739 26334808 15632085 28648823 25401762 26823975 24845553
EMBL
BABH01009997
BABH01009998
BABH01009999
NWSH01002863
PCG67421.1
KQ460036
+ More
KPJ18411.1 PCG67422.1 GDQN01003797 GDQN01001359 JAT87257.1 JAT89695.1 AGBW02013212 OWR43861.1 KQ459593 KPI96422.1 GFDL01010551 JAV24494.1 GFDL01010507 JAV24538.1 GFDF01000980 JAV13104.1 APCN01005391 GFDF01000987 JAV13097.1 AAAB01008898 EAA09144.3 GFDF01000986 JAV13098.1 GFDL01010112 JAV24933.1 AXCM01001884 JXUM01021994 KQ560617 KXJ81601.1 CH477281 EAT44912.1 ATLV01021997 KE525327 KFB47055.1 JXUM01078772 JXUM01078773 KQ563087 KXJ74503.1 KQ434849 KZC08493.1 KQ414637 KOC66978.1 AJWK01005424 AJWK01005425 GL448143 EFN85247.1 KQ766607 OAD53613.1 KQ981153 KYN08902.1 GL887888 EGI69679.1 KQ976424 KYM88410.1 UFQT01000752 SSX26975.1 GGFK01005895 MBW39216.1 GL440368 EFN65983.1 KQ978251 KYM95996.1 LBMM01000070 KMR05083.1 KQ981906 KYN33244.1 GECZ01029064 GECZ01026862 GECZ01006239 GECZ01001712 GECZ01000760 JAS40705.1 JAS42907.1 JAS63530.1 JAS68057.1 JAS69009.1 GL768123 EFZ12082.1 KK107371 EZA51825.1 GAPW01001516 JAC12082.1 GECU01032381 JAS75325.1 JR041408 AEY59436.1 GGFJ01005577 MBW54718.1 GGFM01003161 MBW23912.1 KQ435694 KOX80920.1 SSX26974.1 ADTU01012934 GBYB01006468 JAG76235.1 NEVH01017441 PNF24582.1 CVRI01000021 CRK91685.1 GEBQ01019734 JAT20243.1 DS231898 EDS44609.1 GBYB01006467 JAG76234.1 QOIP01000002 RLU26130.1 KQ982548 KYQ55212.1 CM002910 KMY88986.1 CM000157 EDW88476.1 CH480818 EDW52100.1 GAMC01011282 GAMC01011281 JAB95273.1 AE014134 AY069492 AAF52625.2 AAL39637.1 ADMH02002125 ETN58702.1 CH954177 EDV59322.1 GDKW01000608 JAI55987.1 ACPB03002617 KQ971312 EEZ98053.1 GEZM01033083 JAV84423.1 GFTR01005950 JAW10476.1 CH933807 EDW11175.1 CH963857 EDW75851.2 GBGD01001660 JAC87229.1 CH940649 EDW64096.2 KZ288253 PBC30936.1 OUUW01000004 SPP79785.1 PNF24581.1 CP012523 ALC38200.1 CH379060 EAL33670.1 NNAY01000183 OXU30214.1 GBHO01044042 GBRD01017455 GDHC01019589 JAF99561.1 JAG48372.1 JAP99039.1 KK852561 KDR21339.1 KMY88987.1 AFH03613.1 GECL01002747 JAP03377.1 KRJ97836.1 KQS70752.1 GAMC01011284 GAMC01011283 JAB95272.1
KPJ18411.1 PCG67422.1 GDQN01003797 GDQN01001359 JAT87257.1 JAT89695.1 AGBW02013212 OWR43861.1 KQ459593 KPI96422.1 GFDL01010551 JAV24494.1 GFDL01010507 JAV24538.1 GFDF01000980 JAV13104.1 APCN01005391 GFDF01000987 JAV13097.1 AAAB01008898 EAA09144.3 GFDF01000986 JAV13098.1 GFDL01010112 JAV24933.1 AXCM01001884 JXUM01021994 KQ560617 KXJ81601.1 CH477281 EAT44912.1 ATLV01021997 KE525327 KFB47055.1 JXUM01078772 JXUM01078773 KQ563087 KXJ74503.1 KQ434849 KZC08493.1 KQ414637 KOC66978.1 AJWK01005424 AJWK01005425 GL448143 EFN85247.1 KQ766607 OAD53613.1 KQ981153 KYN08902.1 GL887888 EGI69679.1 KQ976424 KYM88410.1 UFQT01000752 SSX26975.1 GGFK01005895 MBW39216.1 GL440368 EFN65983.1 KQ978251 KYM95996.1 LBMM01000070 KMR05083.1 KQ981906 KYN33244.1 GECZ01029064 GECZ01026862 GECZ01006239 GECZ01001712 GECZ01000760 JAS40705.1 JAS42907.1 JAS63530.1 JAS68057.1 JAS69009.1 GL768123 EFZ12082.1 KK107371 EZA51825.1 GAPW01001516 JAC12082.1 GECU01032381 JAS75325.1 JR041408 AEY59436.1 GGFJ01005577 MBW54718.1 GGFM01003161 MBW23912.1 KQ435694 KOX80920.1 SSX26974.1 ADTU01012934 GBYB01006468 JAG76235.1 NEVH01017441 PNF24582.1 CVRI01000021 CRK91685.1 GEBQ01019734 JAT20243.1 DS231898 EDS44609.1 GBYB01006467 JAG76234.1 QOIP01000002 RLU26130.1 KQ982548 KYQ55212.1 CM002910 KMY88986.1 CM000157 EDW88476.1 CH480818 EDW52100.1 GAMC01011282 GAMC01011281 JAB95273.1 AE014134 AY069492 AAF52625.2 AAL39637.1 ADMH02002125 ETN58702.1 CH954177 EDV59322.1 GDKW01000608 JAI55987.1 ACPB03002617 KQ971312 EEZ98053.1 GEZM01033083 JAV84423.1 GFTR01005950 JAW10476.1 CH933807 EDW11175.1 CH963857 EDW75851.2 GBGD01001660 JAC87229.1 CH940649 EDW64096.2 KZ288253 PBC30936.1 OUUW01000004 SPP79785.1 PNF24581.1 CP012523 ALC38200.1 CH379060 EAL33670.1 NNAY01000183 OXU30214.1 GBHO01044042 GBRD01017455 GDHC01019589 JAF99561.1 JAG48372.1 JAP99039.1 KK852561 KDR21339.1 KMY88987.1 AFH03613.1 GECL01002747 JAP03377.1 KRJ97836.1 KQS70752.1 GAMC01011284 GAMC01011283 JAB95272.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000075840
+ More
UP000075902 UP000076407 UP000075882 UP000007062 UP000075883 UP000075920 UP000075884 UP000075881 UP000075903 UP000075880 UP000075885 UP000069940 UP000249989 UP000008820 UP000030765 UP000076408 UP000076502 UP000075900 UP000053825 UP000092461 UP000008237 UP000078492 UP000007755 UP000078540 UP000000311 UP000078542 UP000036403 UP000078541 UP000053097 UP000069272 UP000053105 UP000005205 UP000235965 UP000183832 UP000002320 UP000279307 UP000002358 UP000075809 UP000002282 UP000001292 UP000000803 UP000000673 UP000008711 UP000015103 UP000007266 UP000192221 UP000009192 UP000007798 UP000008792 UP000242457 UP000268350 UP000005203 UP000092553 UP000001819 UP000215335 UP000027135
UP000075902 UP000076407 UP000075882 UP000007062 UP000075883 UP000075920 UP000075884 UP000075881 UP000075903 UP000075880 UP000075885 UP000069940 UP000249989 UP000008820 UP000030765 UP000076408 UP000076502 UP000075900 UP000053825 UP000092461 UP000008237 UP000078492 UP000007755 UP000078540 UP000000311 UP000078542 UP000036403 UP000078541 UP000053097 UP000069272 UP000053105 UP000005205 UP000235965 UP000183832 UP000002320 UP000279307 UP000002358 UP000075809 UP000002282 UP000001292 UP000000803 UP000000673 UP000008711 UP000015103 UP000007266 UP000192221 UP000009192 UP000007798 UP000008792 UP000242457 UP000268350 UP000005203 UP000092553 UP000001819 UP000215335 UP000027135
Interpro
Gene 3D
ProteinModelPortal
H9JNT2
A0A2A4J630
A0A0N1PHK9
A0A2A4J7N3
A0A1E1WK18
A0A212EQY3
+ More
A0A194PSY3 A0A1Q3FA85 A0A1Q3FAK8 A0A1L8E392 A0A182HYN2 A0A1L8E2Z9 A0A182UCR1 A0A182XIJ6 A0A182KZK3 Q7QA12 A0A1L8E2Z0 A0A1Q3FBQ8 A0A182LV83 A0A182W3F2 A0A182NGE7 A0A182JXK9 A0A182UPX8 A0A182JM65 A0A182PP85 A0A182H072 Q0IG11 A0A1S4F5L1 A0A084WA11 A0A182Y8T2 A0A182GPM3 A0A154P9I3 A0A182RZK6 A0A0L7R820 A0A1B0GHQ2 E2BGD2 A0A310SE47 A0A195D7R9 F4W809 A0A195BPH5 A0A336M9Q2 A0A2M4AEJ7 E2AKQ5 A0A195C748 A0A0J7LB69 A0A195EZE3 A0A1B6EY91 E9J529 A0A026W6Y9 A0A182FSS6 A0A023ES73 A0A087ZJV1 A0A1B6HL57 V9IG08 A0A2M4BPK5 A0A2M3Z5W4 A0A0M9AB75 A0A336M9F1 A0A158NDS6 A0A0C9PY51 A0A2J7Q7M5 A0A1J1HUJ8 A0A1B6L960 B0WDK0 A0A0C9R0L5 A0A3L8E079 K7J5J2 A0A151X475 A0A0J9QYG1 B4NWU6 B4HYH0 W8BE42 Q9VLQ9 W5J2Z9 B3N726 A0A0P4VZH5 T1HLS5 D6WA63 A0A1W4V7T4 A0A1Y1MFI5 A0A224XDD8 B4KGX5 B4MVY1 A0A069DYW9 B4LUA9 A0A2A3EIC4 A0A3B0JCK1 A0A2J7Q7M1 A0A088ATA4 A0A0M4ENG3 Q29MM3 A0A232FHJ3 A0A0A9VTD2 A0A067RNM3 A0A0J9QY14 M9NDI3 A0A0V0G5Q3 A0A0R1DKR4 A0A0Q5WBC1 W8B9F4
A0A194PSY3 A0A1Q3FA85 A0A1Q3FAK8 A0A1L8E392 A0A182HYN2 A0A1L8E2Z9 A0A182UCR1 A0A182XIJ6 A0A182KZK3 Q7QA12 A0A1L8E2Z0 A0A1Q3FBQ8 A0A182LV83 A0A182W3F2 A0A182NGE7 A0A182JXK9 A0A182UPX8 A0A182JM65 A0A182PP85 A0A182H072 Q0IG11 A0A1S4F5L1 A0A084WA11 A0A182Y8T2 A0A182GPM3 A0A154P9I3 A0A182RZK6 A0A0L7R820 A0A1B0GHQ2 E2BGD2 A0A310SE47 A0A195D7R9 F4W809 A0A195BPH5 A0A336M9Q2 A0A2M4AEJ7 E2AKQ5 A0A195C748 A0A0J7LB69 A0A195EZE3 A0A1B6EY91 E9J529 A0A026W6Y9 A0A182FSS6 A0A023ES73 A0A087ZJV1 A0A1B6HL57 V9IG08 A0A2M4BPK5 A0A2M3Z5W4 A0A0M9AB75 A0A336M9F1 A0A158NDS6 A0A0C9PY51 A0A2J7Q7M5 A0A1J1HUJ8 A0A1B6L960 B0WDK0 A0A0C9R0L5 A0A3L8E079 K7J5J2 A0A151X475 A0A0J9QYG1 B4NWU6 B4HYH0 W8BE42 Q9VLQ9 W5J2Z9 B3N726 A0A0P4VZH5 T1HLS5 D6WA63 A0A1W4V7T4 A0A1Y1MFI5 A0A224XDD8 B4KGX5 B4MVY1 A0A069DYW9 B4LUA9 A0A2A3EIC4 A0A3B0JCK1 A0A2J7Q7M1 A0A088ATA4 A0A0M4ENG3 Q29MM3 A0A232FHJ3 A0A0A9VTD2 A0A067RNM3 A0A0J9QY14 M9NDI3 A0A0V0G5Q3 A0A0R1DKR4 A0A0Q5WBC1 W8B9F4
PDB
6E8R
E-value=1.64375e-54,
Score=538
Ontologies
GO
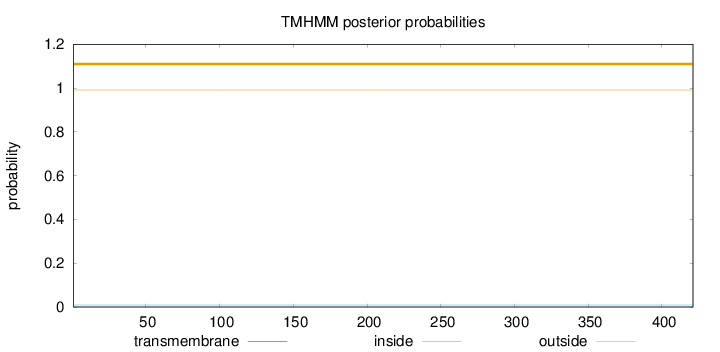
Topology
Length:
421
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00905
outside
1 - 421
Population Genetic Test Statistics
Pi
20.347985
Theta
20.594456
Tajima's D
-1.110404
CLR
26.953514
CSRT
0.11954402279886
Interpretation
Uncertain
