Gene
KWMTBOMO14140
Annotation
endonuclease_and_reverse_transcriptase-like_protein_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.652 PlasmaMembrane Reliability : 1.159
Sequence
CDS
ATGGAGCGAATTATAAATATACAACTCCTGAAGTATCTTGAAGATCGCCAGCTGATCAGTGACCGACAGTACGGTTTCCGTCACGGTCGCTCAGCTGGCGATCTTCTTGTATACCTTACTCACAGGTGGGCTGAAGCCTTGGAGAGCAAGGGCGAGGCTCTTGCTGTGAGCCTTGATATCGCGAAGGCCTTCGACAGGGTCTGGCATAGGGCACTTCTGTCGAAGCTACCATCTTACGGAATCCCCGAGGGTCTCTGCAAGTGGATCGCTAGCTTTTTGGATGGGCGGAGCATCACGGTCGTTGTAGACGGTGACTGTTCTGATACCATGACCATTAACGCTGGCGTTCCACAAGGTTCGGTGCTCTCCCCCACGCTTTTCATCCTGTATATCAATGACATGCTGTCTATTGATGGCATGCATTGCTATGCGGATGACAACACGGGGGATGCGCGATATATCGGCCATCAGAGTCTCTCTCGGAGCGTGGTGCAAGAGAGACGATCAAAACTTGTGTCTGAAGTGGAGAACTCTCTGGGGCGAGTCTCCAAATGGGGTGAATTGAACTTGGTTCAATTCAACCCGTTAAAGACACAAGTTTGCGCGTTCACTGCGAAGAAGGACCCCTTTGTCATGGCGCCGCAATTCCAAGGAGTATCCCTGCAACCTTCCGAGAGTATTGGGATACTTGGGGTCGACATTTCGAGCGATGTCCAGTTTCGGAGTCATTTGGAAGGCAAAGCCAAGTTGGCGTCCAAAATGCTGGGAGTCCTCAACAGAGCGAAGCGGTACTTCACGCCTGGACAAAGGCTTCTACTTTATAAAGAACAAGTCCGGCCTTGCGTGCAGTACTGCTCCCATCTCTGGGCCGGGGCTCCCAAATACCAGCTTCTTTCATTTGACTCCATACAGAGGAGGGCCGTTCGGATTGTCGATAATCCCATTCTCACGGATCGTTTGGAACCTCTGGGTCTGCGGAGGGACTTCGGTTCCCTCTGTATTTTGTACCGTATGTTCCATGGGGAGTGCTCTGAGGAATTGTTCGAGATGATACCGGCATCTCGTTTTTACCATCGCACCGCCCGCCACCGGAGTAGAGTTCATCCGTACTACCTGGAGCCACTGCGGTCATCCACAGTGCGTTTCCAGAGAGTTTTTTTGCCACGTACCATCCGGCTATGGAATGAGCTCCCCTCCACGGTGTTTCCCGAGCGCTATGACATGTTCTTCTTCAAACGAGGCTTGTGGAGAGTATTAAGCGGTAGGCAGCGGCTTGGCTCTGCCTCTGGCATTGCTGAAGTCCATGGGCAACAGTAA
Protein
MERIINIQLLKYLEDRQLISDRQYGFRHGRSAGDLLVYLTHRWAEALESKGEALAVSLDIAKAFDRVWHRALLSKLPSYGIPEGLCKWIASFLDGRSITVVVDGDCSDTMTINAGVPQGSVLSPTLFILYINDMLSIDGMHCYADDNTGDARYIGHQSLSRSVVQERRSKLVSEVENSLGRVSKWGELNLVQFNPLKTQVCAFTAKKDPFVMAPQFQGVSLQPSESIGILGVDISSDVQFRSHLEGKAKLASKMLGVLNRAKRYFTPGQRLLLYKEQVRPCVQYCSHLWAGAPKYQLLSFDSIQRRAVRIVDNPILTDRLEPLGLRRDFGSLCILYRMFHGECSEELFEMIPASRFYHRTARHRSRVHPYYLEPLRSSTVRFQRVFLPRTIRLWNELPSTVFPERYDMFFFKRGLWRVLSGRQRLGSASGIAEVHGQQ
Summary
Cofactor
pyridoxal 5'-phosphate
Uniprot
EMBL
AY359886
AAQ57129.1
RSAL01000002
RVE54942.1
RSAL01000090
RVE48098.1
+ More
RSAL01000422 RVE41792.1 RSAL01002067 RVE40593.1 RSAL01000171 RVE45150.1 DPOC01000269 HCX22453.1 GDRN01105059 JAI57791.1 HAAD01000593 CDG66825.1 GDRN01111334 JAI56796.1 GDRN01111277 JAI56804.1 GEGO01003312 JAR92092.1 GFAC01000919 JAT98269.1 GDRN01111326 JAI56798.1
RSAL01000422 RVE41792.1 RSAL01002067 RVE40593.1 RSAL01000171 RVE45150.1 DPOC01000269 HCX22453.1 GDRN01105059 JAI57791.1 HAAD01000593 CDG66825.1 GDRN01111334 JAI56796.1 GDRN01111277 JAI56804.1 GEGO01003312 JAR92092.1 GFAC01000919 JAT98269.1 GDRN01111326 JAI56798.1
Proteomes
PRIDE
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR007023 Ribosom_reg
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR034653 SPF45_RRM
IPR003954 RRM_dom_euk
IPR000467 G_patch_dom
IPR040052 RBM17
IPR035979 RBD_domain_sf
IPR015424 PyrdxlP-dep_Trfase
IPR002129 PyrdxlP-dep_de-COase
IPR021115 Pyridoxal-P_BS
IPR015421 PyrdxlP-dep_Trfase_major
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR010977 Aromatic_deC
IPR030223 Dispatched
IPR003392 Ptc/Disp
IPR000731 SSD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR007023 Ribosom_reg
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR034653 SPF45_RRM
IPR003954 RRM_dom_euk
IPR000467 G_patch_dom
IPR040052 RBM17
IPR035979 RBD_domain_sf
IPR015424 PyrdxlP-dep_Trfase
IPR002129 PyrdxlP-dep_de-COase
IPR021115 Pyridoxal-P_BS
IPR015421 PyrdxlP-dep_Trfase_major
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR010977 Aromatic_deC
IPR030223 Dispatched
IPR003392 Ptc/Disp
IPR000731 SSD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
Gene 3D
ProteinModelPortal
PDB
6AR3
E-value=8.44181e-05,
Score=110
Ontologies
PANTHER
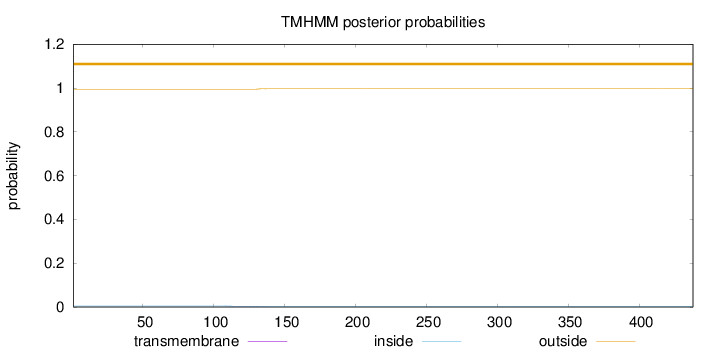
Topology
Length:
438
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04064
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00500
outside
1 - 438
Population Genetic Test Statistics
Pi
0.005786
Theta
0.049506
Tajima's D
-0.485356
CLR
0.45998
CSRT
0.0749462526873656
Interpretation
Uncertain
