Gene
KWMTBOMO14137 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011399
Annotation
AF117598_1_unknown_[Manduca_sexta]
Location in the cell
Extracellular Reliability : 2.812
Sequence
CDS
ATGAAATACCTTATCGTACTGGCCGTTTTCGCTTTGGCGTTCGCCGCTGAAGAGAAGGAGGAGAGGCCGAAGACATTCAGGCGGTTGATTCCAGCTGACGTTTTAAGAGATTTCCCCGGAATGTGCTTCGCGTCCACCCGCTGCATCACCGTCGAGCCCGGCAACACCTGGGAGCTGTCTCCGTTCTGCGGCCGCAGCACCTGCGTCGTCAGCGAGGACCAGCCCCCCAGGCTGCTGGAGCTTGTCGAAGACTGCGGACCTCTGCCACTCACCAACCCCAAGTGCCAATTAGATACCGAAAAGACCAATAAGACCGCTCCCTTCCCTGGCTGCTGCCCGATCTTTACCTGCGAGGAGGGCGCCAAGCTGGAGTACCCTGAGCTCCCCACCCCTCCCCCAGAAGACAAGAAAGCGGAGGAGAAGCCTAAAGCTTAA
Protein
MKYLIVLAVFALAFAAEEKEERPKTFRRLIPADVLRDFPGMCFASTRCITVEPGNTWELSPFCGRSTCVVSEDQPPRLLELVEDCGPLPLTNPKCQLDTEKTNKTAPFPGCCPIFTCEEGAKLEYPELPTPPPEDKKAEEKPKA
Summary
Uniprot
A0A212FBA1
Q9U4Z0
A0A2A4JCY1
A0A2W1BCL8
I4DID5
A0A0N0PE88
+ More
D2A5U4 A0A194PTY7 J3JVX6 A0A1W4XHY5 A0A1L8DAG0 A0A1L8DNQ4 A0A182Q5P9 A0A182NFN3 A0A2M4ATY1 A0A182XW01 A0A182SKK0 A0A182IL13 A0A2M4AT33 A0A182UYN8 A0A182HZY2 A0A0C9QRU1 A0A1D1Z4J4 A0A2M4D109 A0A182MLD4 F5HM67 A0A182VRG9 A0A1B0DCA7 A0A182LR16 A0A1Q3FP50 B0W6P6 A0A1Q3FNZ0 A0A1B0BP77 A0A182GZ09 Q17P92 A0A1A9ZMG6 D3TRB2 A0A088APR8 T1PH12 B4MF17 E2A0E6 B4L7F8 A0A151IFU0 A0A1B6M3L6 E0VL68 A0A1Y1MTV5 A0A195FK05 A0A158NR85 A0A151IU25 A0A151X580 A0A195BFE4 A0A232F6E3 K7ILY3 A0A0A9YPX9 A0A146M2G4 A0A0L7QQB1 A0A2A3E3Q7 A0A1B6JYL6 B4JZR7 F4WRH0 A0A1B6FDH8 A0A2K8JS71 A0A2J7QRL2 A0A1S3CXQ3 A0A2R7VX44 A0A2P8Z4A7 A0A161M4K5 A0A224XSA0 A0A2K8JRG6 A0A0V0GFI2 A0A220XIF5 R4G2U9 A0A069DN82
D2A5U4 A0A194PTY7 J3JVX6 A0A1W4XHY5 A0A1L8DAG0 A0A1L8DNQ4 A0A182Q5P9 A0A182NFN3 A0A2M4ATY1 A0A182XW01 A0A182SKK0 A0A182IL13 A0A2M4AT33 A0A182UYN8 A0A182HZY2 A0A0C9QRU1 A0A1D1Z4J4 A0A2M4D109 A0A182MLD4 F5HM67 A0A182VRG9 A0A1B0DCA7 A0A182LR16 A0A1Q3FP50 B0W6P6 A0A1Q3FNZ0 A0A1B0BP77 A0A182GZ09 Q17P92 A0A1A9ZMG6 D3TRB2 A0A088APR8 T1PH12 B4MF17 E2A0E6 B4L7F8 A0A151IFU0 A0A1B6M3L6 E0VL68 A0A1Y1MTV5 A0A195FK05 A0A158NR85 A0A151IU25 A0A151X580 A0A195BFE4 A0A232F6E3 K7ILY3 A0A0A9YPX9 A0A146M2G4 A0A0L7QQB1 A0A2A3E3Q7 A0A1B6JYL6 B4JZR7 F4WRH0 A0A1B6FDH8 A0A2K8JS71 A0A2J7QRL2 A0A1S3CXQ3 A0A2R7VX44 A0A2P8Z4A7 A0A161M4K5 A0A224XSA0 A0A2K8JRG6 A0A0V0GFI2 A0A220XIF5 R4G2U9 A0A069DN82
Pubmed
EMBL
AGBW02009359
OWR51022.1
AF117598
AAF16720.1
NWSH01002070
PCG69253.1
+ More
KZ150282 PZC71625.1 AK401053 BAM17675.1 KQ460036 KPJ18396.1 KQ971345 EFA05419.1 KQ459593 KPI96438.1 BT127394 AEE62356.1 GFDF01010729 JAV03355.1 GFDF01005981 JAV08103.1 AXCN02000239 GGFK01010944 MBW44265.1 GGFK01010635 MBW43956.1 APCN01002019 GBYB01006449 JAG76216.1 GDJX01006116 JAT61820.1 GGFL01006600 MBW70778.1 AXCM01001191 AAAB01008987 EAA01028.3 EGK97388.1 EGK97389.1 AJVK01030705 GFDL01005777 JAV29268.1 DS231849 EDS36877.1 GFDL01005801 JAV29244.1 JXJN01017834 JXUM01021843 JXUM01021844 JXUM01021845 JXUM01021846 JXUM01021847 KQ560612 KXJ81629.1 CH477193 EAT48540.1 EAT48541.1 CCAG010019338 CCAG010019339 EZ423964 ADD20240.1 KA647425 AFP62054.1 CM000831 CH940665 ACY70485.1 EDW71118.1 GL435586 EFN73083.1 CH933813 EDW10952.2 KQ977771 KYN00012.1 GEBQ01009456 JAT30521.1 DS235271 EEB14124.1 GEZM01027134 JAV86817.1 KQ981522 KYN40592.1 ADTU01023822 ADTU01023823 KQ980970 KYN10923.1 KQ982540 KYQ55420.1 KQ976504 KYM82890.1 NNAY01000803 OXU26426.1 GBHO01009315 JAG34289.1 GDHC01005457 JAQ13172.1 KQ414786 KOC60823.1 KZ288391 PBC26367.1 GECU01003412 JAT04295.1 CH916380 EDV90933.1 GL888285 EGI63241.1 GECZ01021507 JAS48262.1 KY031031 ATU82782.1 NEVH01011891 PNF31231.1 KK854143 PTY12077.1 PYGN01000202 PSN51332.1 GEMB01004088 JAR99174.1 GFTR01002432 JAW13994.1 MF683314 ATU82455.1 GECL01000077 JAP06047.1 MF278679 ASL04985.1 ACPB03016618 GAHY01002276 JAA75234.1 GBGD01003366 JAC85523.1
KZ150282 PZC71625.1 AK401053 BAM17675.1 KQ460036 KPJ18396.1 KQ971345 EFA05419.1 KQ459593 KPI96438.1 BT127394 AEE62356.1 GFDF01010729 JAV03355.1 GFDF01005981 JAV08103.1 AXCN02000239 GGFK01010944 MBW44265.1 GGFK01010635 MBW43956.1 APCN01002019 GBYB01006449 JAG76216.1 GDJX01006116 JAT61820.1 GGFL01006600 MBW70778.1 AXCM01001191 AAAB01008987 EAA01028.3 EGK97388.1 EGK97389.1 AJVK01030705 GFDL01005777 JAV29268.1 DS231849 EDS36877.1 GFDL01005801 JAV29244.1 JXJN01017834 JXUM01021843 JXUM01021844 JXUM01021845 JXUM01021846 JXUM01021847 KQ560612 KXJ81629.1 CH477193 EAT48540.1 EAT48541.1 CCAG010019338 CCAG010019339 EZ423964 ADD20240.1 KA647425 AFP62054.1 CM000831 CH940665 ACY70485.1 EDW71118.1 GL435586 EFN73083.1 CH933813 EDW10952.2 KQ977771 KYN00012.1 GEBQ01009456 JAT30521.1 DS235271 EEB14124.1 GEZM01027134 JAV86817.1 KQ981522 KYN40592.1 ADTU01023822 ADTU01023823 KQ980970 KYN10923.1 KQ982540 KYQ55420.1 KQ976504 KYM82890.1 NNAY01000803 OXU26426.1 GBHO01009315 JAG34289.1 GDHC01005457 JAQ13172.1 KQ414786 KOC60823.1 KZ288391 PBC26367.1 GECU01003412 JAT04295.1 CH916380 EDV90933.1 GL888285 EGI63241.1 GECZ01021507 JAS48262.1 KY031031 ATU82782.1 NEVH01011891 PNF31231.1 KK854143 PTY12077.1 PYGN01000202 PSN51332.1 GEMB01004088 JAR99174.1 GFTR01002432 JAW13994.1 MF683314 ATU82455.1 GECL01000077 JAP06047.1 MF278679 ASL04985.1 ACPB03016618 GAHY01002276 JAA75234.1 GBGD01003366 JAC85523.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000007266
UP000053268
UP000192223
+ More
UP000075886 UP000075884 UP000076408 UP000075901 UP000075880 UP000075903 UP000075840 UP000075883 UP000007062 UP000075920 UP000092462 UP000075882 UP000002320 UP000092460 UP000069940 UP000249989 UP000008820 UP000092445 UP000092444 UP000005203 UP000095301 UP000008792 UP000000311 UP000009192 UP000078542 UP000009046 UP000078541 UP000005205 UP000078492 UP000075809 UP000078540 UP000215335 UP000002358 UP000053825 UP000242457 UP000001070 UP000007755 UP000235965 UP000079169 UP000245037 UP000015103
UP000075886 UP000075884 UP000076408 UP000075901 UP000075880 UP000075903 UP000075840 UP000075883 UP000007062 UP000075920 UP000092462 UP000075882 UP000002320 UP000092460 UP000069940 UP000249989 UP000008820 UP000092445 UP000092444 UP000005203 UP000095301 UP000008792 UP000000311 UP000009192 UP000078542 UP000009046 UP000078541 UP000005205 UP000078492 UP000075809 UP000078540 UP000215335 UP000002358 UP000053825 UP000242457 UP000001070 UP000007755 UP000235965 UP000079169 UP000245037 UP000015103
Pfam
PF15430 SVWC
Interpro
IPR029277
SVWC_dom
ProteinModelPortal
A0A212FBA1
Q9U4Z0
A0A2A4JCY1
A0A2W1BCL8
I4DID5
A0A0N0PE88
+ More
D2A5U4 A0A194PTY7 J3JVX6 A0A1W4XHY5 A0A1L8DAG0 A0A1L8DNQ4 A0A182Q5P9 A0A182NFN3 A0A2M4ATY1 A0A182XW01 A0A182SKK0 A0A182IL13 A0A2M4AT33 A0A182UYN8 A0A182HZY2 A0A0C9QRU1 A0A1D1Z4J4 A0A2M4D109 A0A182MLD4 F5HM67 A0A182VRG9 A0A1B0DCA7 A0A182LR16 A0A1Q3FP50 B0W6P6 A0A1Q3FNZ0 A0A1B0BP77 A0A182GZ09 Q17P92 A0A1A9ZMG6 D3TRB2 A0A088APR8 T1PH12 B4MF17 E2A0E6 B4L7F8 A0A151IFU0 A0A1B6M3L6 E0VL68 A0A1Y1MTV5 A0A195FK05 A0A158NR85 A0A151IU25 A0A151X580 A0A195BFE4 A0A232F6E3 K7ILY3 A0A0A9YPX9 A0A146M2G4 A0A0L7QQB1 A0A2A3E3Q7 A0A1B6JYL6 B4JZR7 F4WRH0 A0A1B6FDH8 A0A2K8JS71 A0A2J7QRL2 A0A1S3CXQ3 A0A2R7VX44 A0A2P8Z4A7 A0A161M4K5 A0A224XSA0 A0A2K8JRG6 A0A0V0GFI2 A0A220XIF5 R4G2U9 A0A069DN82
D2A5U4 A0A194PTY7 J3JVX6 A0A1W4XHY5 A0A1L8DAG0 A0A1L8DNQ4 A0A182Q5P9 A0A182NFN3 A0A2M4ATY1 A0A182XW01 A0A182SKK0 A0A182IL13 A0A2M4AT33 A0A182UYN8 A0A182HZY2 A0A0C9QRU1 A0A1D1Z4J4 A0A2M4D109 A0A182MLD4 F5HM67 A0A182VRG9 A0A1B0DCA7 A0A182LR16 A0A1Q3FP50 B0W6P6 A0A1Q3FNZ0 A0A1B0BP77 A0A182GZ09 Q17P92 A0A1A9ZMG6 D3TRB2 A0A088APR8 T1PH12 B4MF17 E2A0E6 B4L7F8 A0A151IFU0 A0A1B6M3L6 E0VL68 A0A1Y1MTV5 A0A195FK05 A0A158NR85 A0A151IU25 A0A151X580 A0A195BFE4 A0A232F6E3 K7ILY3 A0A0A9YPX9 A0A146M2G4 A0A0L7QQB1 A0A2A3E3Q7 A0A1B6JYL6 B4JZR7 F4WRH0 A0A1B6FDH8 A0A2K8JS71 A0A2J7QRL2 A0A1S3CXQ3 A0A2R7VX44 A0A2P8Z4A7 A0A161M4K5 A0A224XSA0 A0A2K8JRG6 A0A0V0GFI2 A0A220XIF5 R4G2U9 A0A069DN82
Ontologies
GO
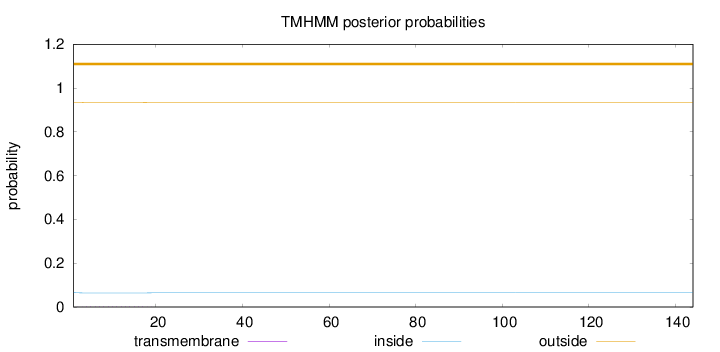
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.998457
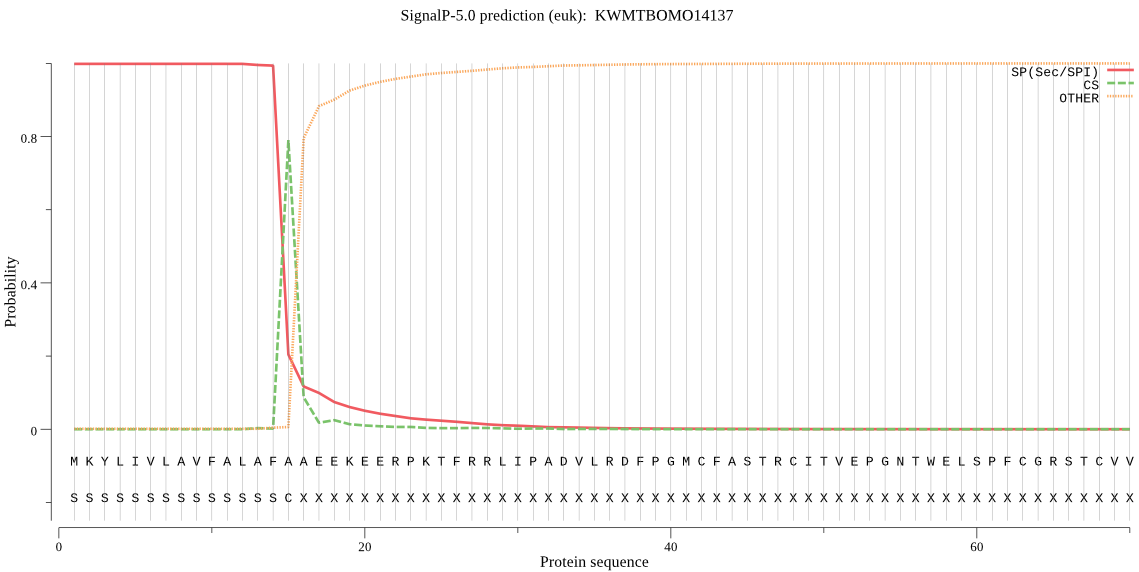
Length:
144
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02823
Exp number, first 60 AAs:
0.02823
Total prob of N-in:
0.06556
outside
1 - 144
Population Genetic Test Statistics
Pi
28.624846
Theta
27.158269
Tajima's D
-1.091864
CLR
1.616561
CSRT
0.11744412779361
Interpretation
Uncertain
