Gene
KWMTBOMO14133
Pre Gene Modal
BGIBMGA011397
Annotation
PREDICTED:_uncharacterized_protein_LOC101741591_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.236
Sequence
CDS
ATGGGCCCGGTCGGAGACAGCTGTTACACAGCTCAGATCGACGCCAGGCGAAATGGAGGACACCAACTACCGGAGAGCCCGCCAGATTCTGGATCGGAGAATCCGTACAGTCCCTCTGAAACCCAGGTGCATACCATAGCCGTACCACAAACGGTTCTCAACGCCGAATACATGCTAGTCCACGAACATTTACCAACCCACGAAATACTCCAACAAAATGGAGATTACATTTACGAGGAGTTGAAATCGGACAACTTGGACCAGGATGTGCTGAGGAGTAATCTGAACGATGTGGTCGTTCTGCCTCAAGATCAGAACTTGGAGTTAGGGATACGGCAGGTTCGCCACGATCTTGGTCTGACCGACTCTGTATACCCGAACAGATTTGGACAGATGAGGGTCGACATGCAAGAGCTGGAGCAAGGCATGATAAGCTCGCAACTGGTATCTCTAGGGCACGAGACTCTGACTCCTGTATATACGAGCCTGCAAGAGCCTAACGTGAAGAAGAGGAAATTATCCCAAGACACAAGTTCTCAAGTTAAATGTGAGCCAGCGGCCTTATCTCCGGAAACAGTGCACCCGGTCAGAGCGCCCGCTTCGCTTGACGGCTCTGAAGCCGGCGACGATGCGCTCTTACAGTGCATCCGCTTCGCATCATTTCAAGAGCGAAGCTGGCATCTTTTGTATGACTGCAATCTGAAGCCACTGCAAACTCCCAGTTACGTGGTCGGAGCCGATAAAGGCTTCAACTATTCTCAAATAGACGATGCTTTCGTCTGCCAGAAGAAGAATCATTTCCAAGTCACCTGCCAGATACAGATGCAGGGAGAACCGAATTACGTCAAAACCGCTGATGGTTTCAAGAAGATCAGTAGCTTTTGTCTACACTTTTACGGAGTGAAGGCCGAAGACCCAAGTCAAAGAGTCAGAGTTGAGCAAAGCCAATCGGATAGAACAAAGAAGCCCTTTCATCCAGTACCAGTGGAGATACGTCGTGAGGGTGCCAAAGTGACAGTCGGGAGACTCCACTTCGCGGAGACCACGAACAACAATATGAGGAAGAAGGGGCGTCCGAACCCTGACCAGAGACACTTCCAGCTGGTCGTGGCCTTGCAAGCCCACGTGGCCAAAGAAGATTACATGCTCATAGCCCATGCCAGTGACAAAATTATTGTCAGGGCGTCGAACCCTGGCCAGTTCGAGTCCGACTGCACGGAGAGCTGGTGGCAACGAGGAGTTGCAGAAAACAGCGTCCACTTCAGCGGGAAGGTCGGCATCAACACGGACAGACCGGACGAGTCCTGCGTCGTCAATGGAAACCTGAAAGTCATGGGCCACATACTCCATCCTTCGGACGAGAGAGCCAAACATAACATAGAAGAGTTAGACACGGCCCAGCAATTGAAAAACGTGCAGAGCATCCGGGTTGTCAAATTCAACTACGACCCTTCATTCGCGGAGCACTCTGGTCTGCTGGGTTACGACCCCGGGCGGCAGGGTCCGCAGCTGGACACCGGAGTCATCGCGCAGGAGGTCCGCCGCGTCATACCAGAGGCGGTCAAGGAGGCCGGTGACGTCACATTGCCCAACGGCGACACCATACCCAAGTTCTTGGTCGTCAATAAGGACCGGATATTCATGGAGAATCTGGGCGCTGTCAAAGAACTGTGCAAGGTGACTGGAAACTTGGAGTCCAGGATAGACCAGCTCGAGAGGATCAACCAGAAGCTGTGCAGGATCACCGTCCTGCAGCGGCGAGGCAGCTCCAGATCAAGCATCAGCAATGACTCCCGATACTCGGCAACATCGAACTCTTCGAAATCGCTCTACAACGACAACGTGTCGATAGATCAGATACGCGACATCGCCAGAAGCATCCGCAGGCACGAGTGTTGTCACAAAATGAACCACAACTCGCCGAGGTACACCAGGAAGCAGTGCAAGCATTGCCACAGCAGCTCCTCAAAATACGGAAAATTTTACAACTCGAATAAGACGTGCTATCGATACAACGATAAGGAGAAGATCGATATGGCCGGCCCTAATTATGTCGATAAAATGTTGCCAGACGACGACTTCGTTGAGTTGCCGAAGAAGAAAGATCACTGTCTATGGGTTAGGAGCGAAGACGATTTCGCGTATGGCAACACTAAATTGGGTTTTTGCTGTCGGAAAGACTACGGGGATGCCAGCAGCGAACTGATCTCCAATAAGTTTTTGCAGATCATCATAACTATATTGGTGTTCGTTATGGCTGTTTGCCTCGTCGTGATATCGGCGCTATACTTCCGGGAGCACCAAGAGCTGCTGACGATGAAGGAGACACGGATGCACGAGAAATTCTCCAGCTACCCTCATTACTCGAAACTGAACGCGGGTAGCGCGCACAAGGCGCCGCCTGACCATAATTTGATACAGAATTTAAAACCGTCCCAACACGTTTTGCCCAAGAAAGTCAAAGAAGGATACATTCTCTAG
Protein
MGPVGDSCYTAQIDARRNGGHQLPESPPDSGSENPYSPSETQVHTIAVPQTVLNAEYMLVHEHLPTHEILQQNGDYIYEELKSDNLDQDVLRSNLNDVVVLPQDQNLELGIRQVRHDLGLTDSVYPNRFGQMRVDMQELEQGMISSQLVSLGHETLTPVYTSLQEPNVKKRKLSQDTSSQVKCEPAALSPETVHPVRAPASLDGSEAGDDALLQCIRFASFQERSWHLLYDCNLKPLQTPSYVVGADKGFNYSQIDDAFVCQKKNHFQVTCQIQMQGEPNYVKTADGFKKISSFCLHFYGVKAEDPSQRVRVEQSQSDRTKKPFHPVPVEIRREGAKVTVGRLHFAETTNNNMRKKGRPNPDQRHFQLVVALQAHVAKEDYMLIAHASDKIIVRASNPGQFESDCTESWWQRGVAENSVHFSGKVGINTDRPDESCVVNGNLKVMGHILHPSDERAKHNIEELDTAQQLKNVQSIRVVKFNYDPSFAEHSGLLGYDPGRQGPQLDTGVIAQEVRRVIPEAVKEAGDVTLPNGDTIPKFLVVNKDRIFMENLGAVKELCKVTGNLESRIDQLERINQKLCRITVLQRRGSSRSSISNDSRYSATSNSSKSLYNDNVSIDQIRDIARSIRRHECCHKMNHNSPRYTRKQCKHCHSSSSKYGKFYNSNKTCYRYNDKEKIDMAGPNYVDKMLPDDDFVELPKKKDHCLWVRSEDDFAYGNTKLGFCCRKDYGDASSELISNKFLQIIITILVFVMAVCLVVISALYFREHQELLTMKETRMHEKFSSYPHYSKLNAGSAHKAPPDHNLIQNLKPSQHVLPKKVKEGYIL
Summary
Uniprot
Pubmed
EMBL
BABH01009953
BABH01009954
BABH01009955
RSAL01000005
RVE54398.1
ODYU01001995
+ More
SOQ38951.1 NWSH01003498 PCG66186.1 KQ459593 KPI96433.1 KQ414620 KOC67683.1 KQ434889 KZC10267.1 GL766836 EFZ14566.1 KQ971344 EFA05214.1 KQ977726 KYN00318.1 GL888217 EGI64695.1 GL438382 EFN69086.1 KZ288285 PBC29504.1 KQ976519 KYM82169.1 KQ981264 KYN44015.1 KQ435755 KOX76053.1 KQ980989 KYN10588.1 KQ982353 KYQ57217.1 ADTU01001338 KK107399 EZA51351.1 QOIP01000002 RLU26124.1 KQ759773 OAD62914.1
SOQ38951.1 NWSH01003498 PCG66186.1 KQ459593 KPI96433.1 KQ414620 KOC67683.1 KQ434889 KZC10267.1 GL766836 EFZ14566.1 KQ971344 EFA05214.1 KQ977726 KYN00318.1 GL888217 EGI64695.1 GL438382 EFN69086.1 KZ288285 PBC29504.1 KQ976519 KYM82169.1 KQ981264 KYN44015.1 KQ435755 KOX76053.1 KQ980989 KYN10588.1 KQ982353 KYQ57217.1 ADTU01001338 KK107399 EZA51351.1 QOIP01000002 RLU26124.1 KQ759773 OAD62914.1
Proteomes
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
PDB
5YHU
E-value=7.15519e-72,
Score=691
Ontologies
GO
PANTHER
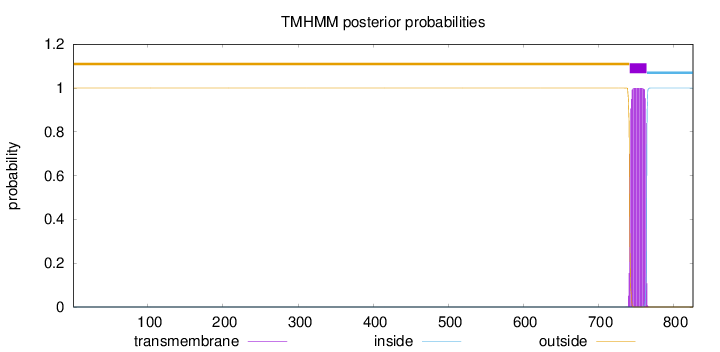
Topology
Length:
826
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.64475
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00005
outside
1 - 741
TMhelix
742 - 764
inside
765 - 826
Population Genetic Test Statistics
Pi
250.540979
Theta
182.782937
Tajima's D
1.233375
CLR
0.668368
CSRT
0.72206389680516
Interpretation
Uncertain
