Gene
KWMTBOMO14128
Pre Gene Modal
BGIBMGA011394
Annotation
PREDICTED:_NADH_dehydrogenase_[ubiquinone]_1_alpha_subcomplex_assembly_factor_5_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.553
Sequence
CDS
ATGATACAAAGAGAAAGAGCAGCCTGTAACGACGAATACCACTTGTCTGAATACGTTAAAGAAGAAATTGGTTGGCGGACAGCTGACAAAATATTCGACATTAAAAGAACGTTCAAAAACGCTGTAGAACTGGGAGCCTCTCGTGGATATGTATCACGGCATTTCCTGCCGGACAGCGTGGAGAAGGTGACACTGTGTGACACATCCAGAACTCATCTCGATAAGGCAATAGTTGGTGAGGGCGTTCAATATGAAAAGATGATAATGGATGAGGAAAATTTTGAATTACCAGACGACAGCGTTGATCTGCTGGTTTCATCGCTTACCTTCCACTGGGTGAACGACCTTCCAGGACTCTTTGACAGGATAATGAAATGCCTCAAACCTGATGGTGTGTTGCTGGCTTGCACCTTCGGTGGAGACACTCTGATGGAGCTGCGTCAGGCGCTCCAACTGTCGGACTTGGAGCGGAGAGGAGGGGTTTCTCCTCATATATCACCGTTTACCAGACCCACGGACATTGGGGGTTTGCTCAATGCTGCTGGTTTTACGTTGCAAACAGTGGACATAGACAAAATGACGATTTGGTACCCCAGTGCCTTTGAGTTGATGCGCGACCTAAGGTTGGCTGGCGAGAGCAACGCAGCCCACAATCGTTCGGGACATCTGCCCCGGGACGTGCAGTTCGCTGCCGCCGCCATATACGACCAGATGTACGGGAAGGACTTCCCCGAGAGGGGTTCCAGGGGAGTCCCGGCTACGTTCGAGATTATCAATTTCGTCGGCTGGAAGCCTGACCCCTCCCAGCCGGCTCCGATCCAGCGCGGCTCGGGACAGGTTTCTCTGAAGGATCTGCACAAAATCGACGAGATATTACAGAAGACAACCACGATTAAAGTTGATGATGATGATAATAAATAA
Protein
MIQRERAACNDEYHLSEYVKEEIGWRTADKIFDIKRTFKNAVELGASRGYVSRHFLPDSVEKVTLCDTSRTHLDKAIVGEGVQYEKMIMDEENFELPDDSVDLLVSSLTFHWVNDLPGLFDRIMKCLKPDGVLLACTFGGDTLMELRQALQLSDLERRGGVSPHISPFTRPTDIGGLLNAAGFTLQTVDIDKMTIWYPSAFELMRDLRLAGESNAAHNRSGHLPRDVQFAAAAIYDQMYGKDFPERGSRGVPATFEIINFVGWKPDPSQPAPIQRGSGQVSLKDLHKIDEILQKTTTIKVDDDDNK
Summary
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Uniprot
A0A194RQQ9
A0A194PS30
A0A2W1BHX1
D6WD45
J3JVM6
A0A1B6C9K2
+ More
A0A1W4W7L8 A0A232EM54 K7J7E5 A0A1Y1LPQ4 A0A1B0BNI9 A0A1A9V8C8 A0A1B0G7Q5 A0A0M5IZN9 B4HQZ6 A0A2P8Z4T6 A0A1A9WIP6 Q7K1S1 B4NNF0 B4P439 A0A1A9Y0P0 B3NRE4 A0A1A9ZWC7 A0A1B6MVE4 B4QEW9 A0A1B6JPU0 A0A3B0J381 A0A1B6FLT8 A0A0L0BSR6 A0A0K8W393 B4H8F9 Q28YM2 A0A0A1X9G4 A0A1W4VQS0 A0A026X4D5 A0A0J7KWQ3 A0A034WIK4 B4JW03 B3MHH1 A0A1B0CIB4 A0A067RDD2 B4KTD2 B4LJD6 A0A1I8P2K3 W8BQ02 W8AZ94 A0A336LU23 A0A2A4IUL0 A0A158P1G5 A0A151X7B1 E9GM67 A0A195F3L8 E2AAD8 A0A2A3E8H7 A0A1B3PDP4 A0A1I8N7C7 A0A182YMI8 F4WYI6 A0A2M4AS22 A0A1Q3FCP2 A0A182SBX1 A0A2M4BTL3 B0XK70 A0A182ITV3 A0A162PKJ2 A0A182THB6 A0A182I5L2 A0A182UP79 Q7QGP3 A0A182X443 A0A182KVH6 W5JTD9 A0A1B3PD92 E9I8Z2 A0A0P6AR91 A0A182PGE4 A0A182Q1L7 A0A2R7WHA4 A0A182FVN6 A0A210PPI2 A0A182GPV6 A0A182MRK2 A0A182RW69 A0A182W2A1 A0A182N347 Q178L8 A0A2S2QTI2 E2C229 A0A023F7U5 A0A2C9KZP5 A0A0P4ZMD1 R4G3N9 A0A2H8TJF2 A0A088A895 A0A2S2PL51 V4AF43 T1JVE0 J9JKK2
A0A1W4W7L8 A0A232EM54 K7J7E5 A0A1Y1LPQ4 A0A1B0BNI9 A0A1A9V8C8 A0A1B0G7Q5 A0A0M5IZN9 B4HQZ6 A0A2P8Z4T6 A0A1A9WIP6 Q7K1S1 B4NNF0 B4P439 A0A1A9Y0P0 B3NRE4 A0A1A9ZWC7 A0A1B6MVE4 B4QEW9 A0A1B6JPU0 A0A3B0J381 A0A1B6FLT8 A0A0L0BSR6 A0A0K8W393 B4H8F9 Q28YM2 A0A0A1X9G4 A0A1W4VQS0 A0A026X4D5 A0A0J7KWQ3 A0A034WIK4 B4JW03 B3MHH1 A0A1B0CIB4 A0A067RDD2 B4KTD2 B4LJD6 A0A1I8P2K3 W8BQ02 W8AZ94 A0A336LU23 A0A2A4IUL0 A0A158P1G5 A0A151X7B1 E9GM67 A0A195F3L8 E2AAD8 A0A2A3E8H7 A0A1B3PDP4 A0A1I8N7C7 A0A182YMI8 F4WYI6 A0A2M4AS22 A0A1Q3FCP2 A0A182SBX1 A0A2M4BTL3 B0XK70 A0A182ITV3 A0A162PKJ2 A0A182THB6 A0A182I5L2 A0A182UP79 Q7QGP3 A0A182X443 A0A182KVH6 W5JTD9 A0A1B3PD92 E9I8Z2 A0A0P6AR91 A0A182PGE4 A0A182Q1L7 A0A2R7WHA4 A0A182FVN6 A0A210PPI2 A0A182GPV6 A0A182MRK2 A0A182RW69 A0A182W2A1 A0A182N347 Q178L8 A0A2S2QTI2 E2C229 A0A023F7U5 A0A2C9KZP5 A0A0P4ZMD1 R4G3N9 A0A2H8TJF2 A0A088A895 A0A2S2PL51 V4AF43 T1JVE0 J9JKK2
Pubmed
26354079
28756777
18362917
19820115
22516182
23537049
+ More
28648823 20075255 28004739 17994087 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 26108605 15632085 25830018 24508170 30249741 25348373 24845553 24495485 21347285 21292972 20798317 25315136 25244985 21719571 12364791 20966253 20920257 23761445 21282665 28812685 26483478 17510324 25474469 15562597 23254933
28648823 20075255 28004739 17994087 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 26108605 15632085 25830018 24508170 30249741 25348373 24845553 24495485 21347285 21292972 20798317 25315136 25244985 21719571 12364791 20966253 20920257 23761445 21282665 28812685 26483478 17510324 25474469 15562597 23254933
EMBL
KQ459875
KPJ19645.1
KQ459593
KPI96246.1
KZ150153
PZC72807.1
+ More
KQ971312 EEZ98317.1 APGK01041016 BT127294 KB740987 KB631964 AEE62256.1 ENN76123.1 ERL87526.1 GEDC01027167 JAS10131.1 NNAY01003420 OXU19439.1 AAZX01000325 GEZM01050304 JAV75623.1 JXJN01017417 CCAG010016890 CP012524 ALC41936.1 CH480816 EDW47793.1 PYGN01000196 PSN51497.1 AE013599 AY061497 AAF58305.1 AAL29045.1 CH964282 EDW85889.1 CM000158 EDW90549.1 CH954179 EDV56096.1 GEBQ01000094 JAT39883.1 CM000362 CM002911 EDX06996.1 KMY93654.1 GECU01006455 JAT01252.1 OUUW01000001 SPP75767.1 GECZ01018601 JAS51168.1 JRES01001422 KNC23023.1 GDHF01006661 JAI45653.1 CH479223 EDW34994.1 CM000071 EAL25943.1 GBXI01006368 JAD07924.1 KK107024 QOIP01000004 EZA62264.1 RLU24015.1 LBMM01002506 KMQ94726.1 GAKP01004800 JAC54152.1 CH916375 EDV98141.1 CH902619 EDV37971.1 AJWK01013177 KK852530 KDR21891.1 CH933808 EDW10644.2 CH940648 EDW60516.2 GAMC01014991 JAB91564.1 GAMC01014993 JAB91562.1 UFQT01000124 SSX20481.1 NWSH01006569 PCG63359.1 ADTU01006541 KQ982450 KYQ56262.1 GL732552 EFX79439.1 KQ981852 KYN34981.1 GL438095 EFN69604.1 KZ288325 PBC28025.1 KU660030 AOG17828.1 GL888450 EGI60718.1 GGFK01010211 MBW43532.1 GFDL01009709 JAV25336.1 GGFJ01007285 MBW56426.1 DS233727 EDS31377.1 LRGB01000494 KZS18926.1 APCN01003459 AAAB01008823 EAA05526.3 ADMH02000489 ETN66220.1 KU659906 AOG17705.1 GL761667 EFZ22937.1 GDIP01031697 JAM72018.1 AXCN02001125 KK854816 PTY18996.1 NEDP02005568 OWF38377.1 JXUM01016533 KQ560461 KXJ82301.1 AXCM01003706 CH477362 EAT42624.1 GGMS01011841 MBY81044.1 GL452067 EFN77980.1 GBBI01001397 JAC17315.1 GDIP01210485 JAJ12917.1 GAHY01002060 JAA75450.1 GFXV01002316 MBW14121.1 GGMR01017550 MBY30169.1 KB202163 ESO91946.1 CAEY01000792 ABLF02038296
KQ971312 EEZ98317.1 APGK01041016 BT127294 KB740987 KB631964 AEE62256.1 ENN76123.1 ERL87526.1 GEDC01027167 JAS10131.1 NNAY01003420 OXU19439.1 AAZX01000325 GEZM01050304 JAV75623.1 JXJN01017417 CCAG010016890 CP012524 ALC41936.1 CH480816 EDW47793.1 PYGN01000196 PSN51497.1 AE013599 AY061497 AAF58305.1 AAL29045.1 CH964282 EDW85889.1 CM000158 EDW90549.1 CH954179 EDV56096.1 GEBQ01000094 JAT39883.1 CM000362 CM002911 EDX06996.1 KMY93654.1 GECU01006455 JAT01252.1 OUUW01000001 SPP75767.1 GECZ01018601 JAS51168.1 JRES01001422 KNC23023.1 GDHF01006661 JAI45653.1 CH479223 EDW34994.1 CM000071 EAL25943.1 GBXI01006368 JAD07924.1 KK107024 QOIP01000004 EZA62264.1 RLU24015.1 LBMM01002506 KMQ94726.1 GAKP01004800 JAC54152.1 CH916375 EDV98141.1 CH902619 EDV37971.1 AJWK01013177 KK852530 KDR21891.1 CH933808 EDW10644.2 CH940648 EDW60516.2 GAMC01014991 JAB91564.1 GAMC01014993 JAB91562.1 UFQT01000124 SSX20481.1 NWSH01006569 PCG63359.1 ADTU01006541 KQ982450 KYQ56262.1 GL732552 EFX79439.1 KQ981852 KYN34981.1 GL438095 EFN69604.1 KZ288325 PBC28025.1 KU660030 AOG17828.1 GL888450 EGI60718.1 GGFK01010211 MBW43532.1 GFDL01009709 JAV25336.1 GGFJ01007285 MBW56426.1 DS233727 EDS31377.1 LRGB01000494 KZS18926.1 APCN01003459 AAAB01008823 EAA05526.3 ADMH02000489 ETN66220.1 KU659906 AOG17705.1 GL761667 EFZ22937.1 GDIP01031697 JAM72018.1 AXCN02001125 KK854816 PTY18996.1 NEDP02005568 OWF38377.1 JXUM01016533 KQ560461 KXJ82301.1 AXCM01003706 CH477362 EAT42624.1 GGMS01011841 MBY81044.1 GL452067 EFN77980.1 GBBI01001397 JAC17315.1 GDIP01210485 JAJ12917.1 GAHY01002060 JAA75450.1 GFXV01002316 MBW14121.1 GGMR01017550 MBY30169.1 KB202163 ESO91946.1 CAEY01000792 ABLF02038296
Proteomes
UP000053240
UP000053268
UP000007266
UP000019118
UP000030742
UP000192223
+ More
UP000215335 UP000002358 UP000092460 UP000078200 UP000092444 UP000092553 UP000001292 UP000245037 UP000091820 UP000000803 UP000007798 UP000002282 UP000092443 UP000008711 UP000092445 UP000000304 UP000268350 UP000037069 UP000008744 UP000001819 UP000192221 UP000053097 UP000279307 UP000036403 UP000001070 UP000007801 UP000092461 UP000027135 UP000009192 UP000008792 UP000095300 UP000218220 UP000005205 UP000075809 UP000000305 UP000078541 UP000000311 UP000242457 UP000095301 UP000076408 UP000007755 UP000075901 UP000002320 UP000075880 UP000076858 UP000075902 UP000075840 UP000075903 UP000007062 UP000076407 UP000075882 UP000000673 UP000075885 UP000075886 UP000069272 UP000242188 UP000069940 UP000249989 UP000075883 UP000075900 UP000075920 UP000075884 UP000008820 UP000008237 UP000076420 UP000005203 UP000030746 UP000015104 UP000007819
UP000215335 UP000002358 UP000092460 UP000078200 UP000092444 UP000092553 UP000001292 UP000245037 UP000091820 UP000000803 UP000007798 UP000002282 UP000092443 UP000008711 UP000092445 UP000000304 UP000268350 UP000037069 UP000008744 UP000001819 UP000192221 UP000053097 UP000279307 UP000036403 UP000001070 UP000007801 UP000092461 UP000027135 UP000009192 UP000008792 UP000095300 UP000218220 UP000005205 UP000075809 UP000000305 UP000078541 UP000000311 UP000242457 UP000095301 UP000076408 UP000007755 UP000075901 UP000002320 UP000075880 UP000076858 UP000075902 UP000075840 UP000075903 UP000007062 UP000076407 UP000075882 UP000000673 UP000075885 UP000075886 UP000069272 UP000242188 UP000069940 UP000249989 UP000075883 UP000075900 UP000075920 UP000075884 UP000008820 UP000008237 UP000076420 UP000005203 UP000030746 UP000015104 UP000007819
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A194RQQ9
A0A194PS30
A0A2W1BHX1
D6WD45
J3JVM6
A0A1B6C9K2
+ More
A0A1W4W7L8 A0A232EM54 K7J7E5 A0A1Y1LPQ4 A0A1B0BNI9 A0A1A9V8C8 A0A1B0G7Q5 A0A0M5IZN9 B4HQZ6 A0A2P8Z4T6 A0A1A9WIP6 Q7K1S1 B4NNF0 B4P439 A0A1A9Y0P0 B3NRE4 A0A1A9ZWC7 A0A1B6MVE4 B4QEW9 A0A1B6JPU0 A0A3B0J381 A0A1B6FLT8 A0A0L0BSR6 A0A0K8W393 B4H8F9 Q28YM2 A0A0A1X9G4 A0A1W4VQS0 A0A026X4D5 A0A0J7KWQ3 A0A034WIK4 B4JW03 B3MHH1 A0A1B0CIB4 A0A067RDD2 B4KTD2 B4LJD6 A0A1I8P2K3 W8BQ02 W8AZ94 A0A336LU23 A0A2A4IUL0 A0A158P1G5 A0A151X7B1 E9GM67 A0A195F3L8 E2AAD8 A0A2A3E8H7 A0A1B3PDP4 A0A1I8N7C7 A0A182YMI8 F4WYI6 A0A2M4AS22 A0A1Q3FCP2 A0A182SBX1 A0A2M4BTL3 B0XK70 A0A182ITV3 A0A162PKJ2 A0A182THB6 A0A182I5L2 A0A182UP79 Q7QGP3 A0A182X443 A0A182KVH6 W5JTD9 A0A1B3PD92 E9I8Z2 A0A0P6AR91 A0A182PGE4 A0A182Q1L7 A0A2R7WHA4 A0A182FVN6 A0A210PPI2 A0A182GPV6 A0A182MRK2 A0A182RW69 A0A182W2A1 A0A182N347 Q178L8 A0A2S2QTI2 E2C229 A0A023F7U5 A0A2C9KZP5 A0A0P4ZMD1 R4G3N9 A0A2H8TJF2 A0A088A895 A0A2S2PL51 V4AF43 T1JVE0 J9JKK2
A0A1W4W7L8 A0A232EM54 K7J7E5 A0A1Y1LPQ4 A0A1B0BNI9 A0A1A9V8C8 A0A1B0G7Q5 A0A0M5IZN9 B4HQZ6 A0A2P8Z4T6 A0A1A9WIP6 Q7K1S1 B4NNF0 B4P439 A0A1A9Y0P0 B3NRE4 A0A1A9ZWC7 A0A1B6MVE4 B4QEW9 A0A1B6JPU0 A0A3B0J381 A0A1B6FLT8 A0A0L0BSR6 A0A0K8W393 B4H8F9 Q28YM2 A0A0A1X9G4 A0A1W4VQS0 A0A026X4D5 A0A0J7KWQ3 A0A034WIK4 B4JW03 B3MHH1 A0A1B0CIB4 A0A067RDD2 B4KTD2 B4LJD6 A0A1I8P2K3 W8BQ02 W8AZ94 A0A336LU23 A0A2A4IUL0 A0A158P1G5 A0A151X7B1 E9GM67 A0A195F3L8 E2AAD8 A0A2A3E8H7 A0A1B3PDP4 A0A1I8N7C7 A0A182YMI8 F4WYI6 A0A2M4AS22 A0A1Q3FCP2 A0A182SBX1 A0A2M4BTL3 B0XK70 A0A182ITV3 A0A162PKJ2 A0A182THB6 A0A182I5L2 A0A182UP79 Q7QGP3 A0A182X443 A0A182KVH6 W5JTD9 A0A1B3PD92 E9I8Z2 A0A0P6AR91 A0A182PGE4 A0A182Q1L7 A0A2R7WHA4 A0A182FVN6 A0A210PPI2 A0A182GPV6 A0A182MRK2 A0A182RW69 A0A182W2A1 A0A182N347 Q178L8 A0A2S2QTI2 E2C229 A0A023F7U5 A0A2C9KZP5 A0A0P4ZMD1 R4G3N9 A0A2H8TJF2 A0A088A895 A0A2S2PL51 V4AF43 T1JVE0 J9JKK2
PDB
3BKW
E-value=0.000201733,
Score=105
Ontologies
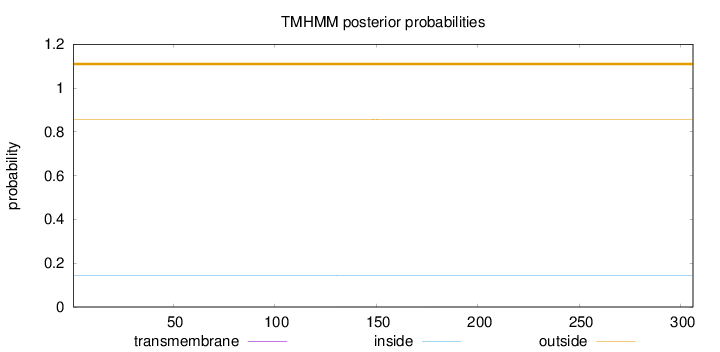
Topology
Length:
306
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01757
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14437
outside
1 - 306
Population Genetic Test Statistics
Pi
259.901061
Theta
172.852851
Tajima's D
1.427673
CLR
0
CSRT
0.763561821908905
Interpretation
Uncertain
