Gene
KWMTBOMO14125
Annotation
PREDICTED:_uncharacterized_protein_LOC101741223_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.044 Nuclear Reliability : 2.511
Sequence
CDS
ATGACTTCGTCTCTTTGGCATGAACTCGTCCCCTCTGCTAATTACAGAAATGAGATTAAGAATGCTCCGGCGACAGGTGGCGGCTGCACGTTTCCAGAGGGCTGGACGGGTAGCTGGTTCCAGTCCGGGAATCCGGGTCTAATCACAATCAACAAAACCCATATACAGATGAAGGGAGAATGCTATGAGACAGACTCCAATGATAAATTCCTTTTGTATTACAGAAATGAGGACTGCTACAGATGCATGGTGATACACGAGAAACACAAATACGTCTTACAGTACAAAGAGACATACTGCAACTCCAAGGACTCTTTGTCGTCAATGTGCGAGTCGATCACCGGGGACGCGTCGCTCTACTCCATGTTCAGAAAGGAGCCAAAGCCTGTGCCGCAGCCCTGCCCCTTCCACCCGGCGCCCTTCACCTTCACATATTATCGAGGCTCTGAAGAGTGCACGTACCCATTGTCCCGTGCCGAATCGTGTACTGACGATTCGCGGCTGCTGCTCCGGTATATGGCGTGCGCCGATATGCTCAGCTCGGAGAGCAACGTTGAAGAGCTGGTGTGCCTAGCTACATGGAAGGAAGGTTCCACCAGGTACCTGGTGGGGCAAATCTCGCAGGTTCAGCGGAGGAATTCTATAGCTTCGGATGAAGACACTTATCGATGTTTCATCTACAAAGGACAGCACACGGATAAAACGACGACATACACGATAGCCCAGTCCGCGGACGCGACCTGCAACGGTCTCTCCTCACCGATGGACGGAAGTCGAACCATGAAGTTGACCAGCAGTGACGACGAACACAACCGCTGCCATTTCCCGAGCTGGATCGTGCAGCACCACAAATGGTTCAGTCTGGACCACACGCACCAGTACCATTTCACAACCAAAAACGCTACCCTCAAGATAATCAACGAGGACGGTTCGTTTGAGGAGCGCAGGCTAGTGTGCCACTCTATACTCGAGCAGAAGGACAAGAAGTATATTAAGCTTGTTGCACACGTCACGAAGGGGTGTGAGTCTGGTCACCAGTGCGTGCAGTTCCACCGGCGCGAGTCCTCCGTGCTGGAGCTCCAGGCGGGCGGGCAGCTAGCGGACGCGCCGCACGACGCATGCGCGCCCCCCCACCTCGCCGCGCCACACGTCACCCTCATCACAACTTCATTGACCCCCACCCGCTGCCCCATTCTCGGGCGGTATTCCGTGGTCAGTTCCTTGGCTGACAGACGGAGAAGGCGGCAGCAGCTCACCATAGGTGATGAAGAGGCGGACGAGCAGCGCGCGGCGGAGTGCGTCGTCGGCGACTACCGGAGTGCCAGCCTCGGCTGCGGGCACGCGCAGGACTCCATCGAGTTCCGTTCACCATGCTCCAGCACAGTGTATACATGTCACGGAGCGTGGCAGGCCGGCTCGAAGGGGTTCGTGCTGGTGTACGACGCGCGCGCCGTCCGGCCCCGCGCTTCCTGCCTCGTGTACACTGCGCCGTCCACCAACGACACGCGCGCGGGCACGGTGCAGCAGCTGTCGCTGAGCGTGATGTCCCACTCGTGCGACCGCTCCGTGCAGCCGGGCGTCGCCGGGGACAAGGCCTACAACCTAACATTAAACGGAACTTGCGAACAAAGCAGCAAATATAGCGCCGCCGTCAGCCCTGCGCCGGCGGTCTTCGTTTTGATGACGTCACTGGCCACGCTTCCCCTCAGATGA
Protein
MTSSLWHELVPSANYRNEIKNAPATGGGCTFPEGWTGSWFQSGNPGLITINKTHIQMKGECYETDSNDKFLLYYRNEDCYRCMVIHEKHKYVLQYKETYCNSKDSLSSMCESITGDASLYSMFRKEPKPVPQPCPFHPAPFTFTYYRGSEECTYPLSRAESCTDDSRLLLRYMACADMLSSESNVEELVCLATWKEGSTRYLVGQISQVQRRNSIASDEDTYRCFIYKGQHTDKTTTYTIAQSADATCNGLSSPMDGSRTMKLTSSDDEHNRCHFPSWIVQHHKWFSLDHTHQYHFTTKNATLKIINEDGSFEERRLVCHSILEQKDKKYIKLVAHVTKGCESGHQCVQFHRRESSVLELQAGGQLADAPHDACAPPHLAAPHVTLITTSLTPTRCPILGRYSVVSSLADRRRRRQQLTIGDEEADEQRAAECVVGDYRSASLGCGHAQDSIEFRSPCSSTVYTCHGAWQAGSKGFVLVYDARAVRPRASCLVYTAPSTNDTRAGTVQQLSLSVMSHSCDRSVQPGVAGDKAYNLTLNGTCEQSSKYSAAVSPAPAVFVLMTSLATLPLR
Summary
Uniprot
A0A2W1BFR1
A0A2A4JWV5
A0A194RP66
A0A212F4N6
A0A194PSF0
H9JNT8
+ More
A0A1Y1KXV5 A0A2J7PZU0 A0A0T6B809 A0A1B6BXW0 A0A1B6EC96 E0VL77 A0A1B6MU61 A0A2K8JMZ5 A0A1B6G0D0 A0A139WBC4 A0A1B6D3A2 A0A067R8C5 A0A2P8Y522 A0A1B6MNT4 A0A0C9QCI9 A0A1B6KQ90 A0A0C9PJ82 A0A1Y1M8S9 A0A2K8JX84 A0A1Y1M9F2 A0A224XGN8 A0A2S2QJW7 A0A195DFD9 A0A026WS51 A0A2S2PLC0 F4W5T6 A0A151X2X5 J9K6E3 E2AKQ1 A0A195BT20 E2BHA9 T1JKC5 A0A158P2S3 A0A0J7P184 A0A0N0U7Z9 T1IAU8 A0A1B6ML19 A0A0A9XH85 A0A0L7RAY6 A0A212EWH7 A0A2H1X2M0 A0A195EZK7 K7JPA1 A0A195C0Q2 A0A154PA97 A0A194REJ8 A0A226D6W0 L7MBC5 A0A224Y7V5 A0A131YWB4 A0A1W7RAD8 A0A1B6D8I4 A0A154P9F6 A0A1E1X264 A0A195CSX1 A0A2S2Q4U0 A0A2S2NU45 A0A158NND8 A0A2R7WNZ2 A0A195FWU2 E9I8G0 A0A0P4YNL4 J9M457 A0A0L7QVX6 A0A1B6GVM4 A0A0P6I4F7 K7J8V1 A0A1Q3FZ85 A0A226D5L0 A0A1S4F452 A0A131Y5K2 A0A0P5K7U8
A0A1Y1KXV5 A0A2J7PZU0 A0A0T6B809 A0A1B6BXW0 A0A1B6EC96 E0VL77 A0A1B6MU61 A0A2K8JMZ5 A0A1B6G0D0 A0A139WBC4 A0A1B6D3A2 A0A067R8C5 A0A2P8Y522 A0A1B6MNT4 A0A0C9QCI9 A0A1B6KQ90 A0A0C9PJ82 A0A1Y1M8S9 A0A2K8JX84 A0A1Y1M9F2 A0A224XGN8 A0A2S2QJW7 A0A195DFD9 A0A026WS51 A0A2S2PLC0 F4W5T6 A0A151X2X5 J9K6E3 E2AKQ1 A0A195BT20 E2BHA9 T1JKC5 A0A158P2S3 A0A0J7P184 A0A0N0U7Z9 T1IAU8 A0A1B6ML19 A0A0A9XH85 A0A0L7RAY6 A0A212EWH7 A0A2H1X2M0 A0A195EZK7 K7JPA1 A0A195C0Q2 A0A154PA97 A0A194REJ8 A0A226D6W0 L7MBC5 A0A224Y7V5 A0A131YWB4 A0A1W7RAD8 A0A1B6D8I4 A0A154P9F6 A0A1E1X264 A0A195CSX1 A0A2S2Q4U0 A0A2S2NU45 A0A158NND8 A0A2R7WNZ2 A0A195FWU2 E9I8G0 A0A0P4YNL4 J9M457 A0A0L7QVX6 A0A1B6GVM4 A0A0P6I4F7 K7J8V1 A0A1Q3FZ85 A0A226D5L0 A0A1S4F452 A0A131Y5K2 A0A0P5K7U8
Pubmed
EMBL
KZ150286
PZC71610.1
NWSH01000477
PCG76124.1
KQ459875
KPJ19638.1
+ More
AGBW02010328 OWR48708.1 KQ459593 KPI96237.1 BABH01009925 BABH01009926 BABH01009927 GEZM01070896 JAV66229.1 NEVH01020332 PNF21842.1 LJIG01009230 KRT83496.1 GEDC01031171 JAS06127.1 GEDC01001732 JAS35566.1 DS235271 EEB14133.1 GEBQ01000532 JAT39445.1 KY031290 ATU83041.1 GECZ01013871 JAS55898.1 KQ971372 KYB25227.1 GEDC01017109 JAS20189.1 KK852869 KDR14671.1 PYGN01000918 PSN39357.1 GEBQ01002373 JAT37604.1 GBYB01001019 JAG70786.1 GEBQ01029719 GEBQ01026593 JAT10258.1 JAT13384.1 GBYB01001018 JAG70785.1 GEZM01037084 JAV82202.1 KY031276 ATU83027.1 GEZM01037085 JAV82201.1 GFTR01007448 JAW08978.1 GGMS01008269 MBY77472.1 KQ980903 KYN11610.1 KK107138 QOIP01000002 EZA57924.1 RLU25641.1 GGMR01017586 MBY30205.1 GL887695 EGI70412.1 KQ982576 KYQ54649.1 ABLF02026618 ABLF02026629 GL440368 EFN65991.1 KQ976417 KYM89584.1 GL448287 EFN84878.1 JH432024 ADTU01007429 LBMM01000418 KMQ98415.1 KQ435694 KOX80982.1 ACPB03000859 GEBQ01003327 JAT36650.1 GBHO01025466 GBRD01006672 GDHC01016466 JAG18138.1 JAG59149.1 JAQ02163.1 KQ414617 KOC68029.1 AGBW02011989 OWR45836.1 ODYU01012522 SOQ58884.1 KQ981905 KYN33347.1 AAZX01003820 AAZX01006549 AAZX01013727 AAZX01021517 KQ978379 KYM94469.1 KQ434856 KZC08763.1 KQ460323 KPJ15889.1 LNIX01000033 OXA40598.1 GACK01004575 JAA60459.1 GFPF01002562 MAA13708.1 GEDV01006101 JAP82456.1 GFAH01000280 JAV48109.1 GEDC01015326 JAS21972.1 KQ434832 KZC07858.1 GFAC01005880 JAT93308.1 KQ977372 KYN03224.1 GGMS01003533 MBY72736.1 GGMR01008124 MBY20743.1 ADTU01021279 ADTU01021280 ADTU01021281 ADTU01021282 ADTU01021283 ADTU01021284 ADTU01021285 ADTU01021286 ADTU01021287 KK855186 PTY21348.1 KQ981208 KYN44787.1 GL761538 EFZ23212.1 GDIP01224676 JAI98725.1 ABLF02039025 ABLF02039034 KQ414721 KOC62783.1 GECZ01003396 JAS66373.1 GDIQ01009473 JAN85264.1 AAZX01000810 AAZX01004306 AAZX01004505 AAZX01005770 AAZX01011008 GFDL01002195 JAV32850.1 OXA40499.1 GEFM01002031 JAP73765.1 GDIQ01189249 JAK62476.1
AGBW02010328 OWR48708.1 KQ459593 KPI96237.1 BABH01009925 BABH01009926 BABH01009927 GEZM01070896 JAV66229.1 NEVH01020332 PNF21842.1 LJIG01009230 KRT83496.1 GEDC01031171 JAS06127.1 GEDC01001732 JAS35566.1 DS235271 EEB14133.1 GEBQ01000532 JAT39445.1 KY031290 ATU83041.1 GECZ01013871 JAS55898.1 KQ971372 KYB25227.1 GEDC01017109 JAS20189.1 KK852869 KDR14671.1 PYGN01000918 PSN39357.1 GEBQ01002373 JAT37604.1 GBYB01001019 JAG70786.1 GEBQ01029719 GEBQ01026593 JAT10258.1 JAT13384.1 GBYB01001018 JAG70785.1 GEZM01037084 JAV82202.1 KY031276 ATU83027.1 GEZM01037085 JAV82201.1 GFTR01007448 JAW08978.1 GGMS01008269 MBY77472.1 KQ980903 KYN11610.1 KK107138 QOIP01000002 EZA57924.1 RLU25641.1 GGMR01017586 MBY30205.1 GL887695 EGI70412.1 KQ982576 KYQ54649.1 ABLF02026618 ABLF02026629 GL440368 EFN65991.1 KQ976417 KYM89584.1 GL448287 EFN84878.1 JH432024 ADTU01007429 LBMM01000418 KMQ98415.1 KQ435694 KOX80982.1 ACPB03000859 GEBQ01003327 JAT36650.1 GBHO01025466 GBRD01006672 GDHC01016466 JAG18138.1 JAG59149.1 JAQ02163.1 KQ414617 KOC68029.1 AGBW02011989 OWR45836.1 ODYU01012522 SOQ58884.1 KQ981905 KYN33347.1 AAZX01003820 AAZX01006549 AAZX01013727 AAZX01021517 KQ978379 KYM94469.1 KQ434856 KZC08763.1 KQ460323 KPJ15889.1 LNIX01000033 OXA40598.1 GACK01004575 JAA60459.1 GFPF01002562 MAA13708.1 GEDV01006101 JAP82456.1 GFAH01000280 JAV48109.1 GEDC01015326 JAS21972.1 KQ434832 KZC07858.1 GFAC01005880 JAT93308.1 KQ977372 KYN03224.1 GGMS01003533 MBY72736.1 GGMR01008124 MBY20743.1 ADTU01021279 ADTU01021280 ADTU01021281 ADTU01021282 ADTU01021283 ADTU01021284 ADTU01021285 ADTU01021286 ADTU01021287 KK855186 PTY21348.1 KQ981208 KYN44787.1 GL761538 EFZ23212.1 GDIP01224676 JAI98725.1 ABLF02039025 ABLF02039034 KQ414721 KOC62783.1 GECZ01003396 JAS66373.1 GDIQ01009473 JAN85264.1 AAZX01000810 AAZX01004306 AAZX01004505 AAZX01005770 AAZX01011008 GFDL01002195 JAV32850.1 OXA40499.1 GEFM01002031 JAP73765.1 GDIQ01189249 JAK62476.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000005204
UP000235965
+ More
UP000009046 UP000007266 UP000027135 UP000245037 UP000078492 UP000053097 UP000279307 UP000007755 UP000075809 UP000007819 UP000000311 UP000078540 UP000008237 UP000005205 UP000036403 UP000053105 UP000015103 UP000053825 UP000078541 UP000002358 UP000078542 UP000076502 UP000198287
UP000009046 UP000007266 UP000027135 UP000245037 UP000078492 UP000053097 UP000279307 UP000007755 UP000075809 UP000007819 UP000000311 UP000078540 UP000008237 UP000005205 UP000036403 UP000053105 UP000015103 UP000053825 UP000078541 UP000002358 UP000078542 UP000076502 UP000198287
PRIDE
Interpro
SUPFAM
SSF52980
SSF52980
Gene 3D
ProteinModelPortal
A0A2W1BFR1
A0A2A4JWV5
A0A194RP66
A0A212F4N6
A0A194PSF0
H9JNT8
+ More
A0A1Y1KXV5 A0A2J7PZU0 A0A0T6B809 A0A1B6BXW0 A0A1B6EC96 E0VL77 A0A1B6MU61 A0A2K8JMZ5 A0A1B6G0D0 A0A139WBC4 A0A1B6D3A2 A0A067R8C5 A0A2P8Y522 A0A1B6MNT4 A0A0C9QCI9 A0A1B6KQ90 A0A0C9PJ82 A0A1Y1M8S9 A0A2K8JX84 A0A1Y1M9F2 A0A224XGN8 A0A2S2QJW7 A0A195DFD9 A0A026WS51 A0A2S2PLC0 F4W5T6 A0A151X2X5 J9K6E3 E2AKQ1 A0A195BT20 E2BHA9 T1JKC5 A0A158P2S3 A0A0J7P184 A0A0N0U7Z9 T1IAU8 A0A1B6ML19 A0A0A9XH85 A0A0L7RAY6 A0A212EWH7 A0A2H1X2M0 A0A195EZK7 K7JPA1 A0A195C0Q2 A0A154PA97 A0A194REJ8 A0A226D6W0 L7MBC5 A0A224Y7V5 A0A131YWB4 A0A1W7RAD8 A0A1B6D8I4 A0A154P9F6 A0A1E1X264 A0A195CSX1 A0A2S2Q4U0 A0A2S2NU45 A0A158NND8 A0A2R7WNZ2 A0A195FWU2 E9I8G0 A0A0P4YNL4 J9M457 A0A0L7QVX6 A0A1B6GVM4 A0A0P6I4F7 K7J8V1 A0A1Q3FZ85 A0A226D5L0 A0A1S4F452 A0A131Y5K2 A0A0P5K7U8
A0A1Y1KXV5 A0A2J7PZU0 A0A0T6B809 A0A1B6BXW0 A0A1B6EC96 E0VL77 A0A1B6MU61 A0A2K8JMZ5 A0A1B6G0D0 A0A139WBC4 A0A1B6D3A2 A0A067R8C5 A0A2P8Y522 A0A1B6MNT4 A0A0C9QCI9 A0A1B6KQ90 A0A0C9PJ82 A0A1Y1M8S9 A0A2K8JX84 A0A1Y1M9F2 A0A224XGN8 A0A2S2QJW7 A0A195DFD9 A0A026WS51 A0A2S2PLC0 F4W5T6 A0A151X2X5 J9K6E3 E2AKQ1 A0A195BT20 E2BHA9 T1JKC5 A0A158P2S3 A0A0J7P184 A0A0N0U7Z9 T1IAU8 A0A1B6ML19 A0A0A9XH85 A0A0L7RAY6 A0A212EWH7 A0A2H1X2M0 A0A195EZK7 K7JPA1 A0A195C0Q2 A0A154PA97 A0A194REJ8 A0A226D6W0 L7MBC5 A0A224Y7V5 A0A131YWB4 A0A1W7RAD8 A0A1B6D8I4 A0A154P9F6 A0A1E1X264 A0A195CSX1 A0A2S2Q4U0 A0A2S2NU45 A0A158NND8 A0A2R7WNZ2 A0A195FWU2 E9I8G0 A0A0P4YNL4 J9M457 A0A0L7QVX6 A0A1B6GVM4 A0A0P6I4F7 K7J8V1 A0A1Q3FZ85 A0A226D5L0 A0A1S4F452 A0A131Y5K2 A0A0P5K7U8
Ontologies
GO
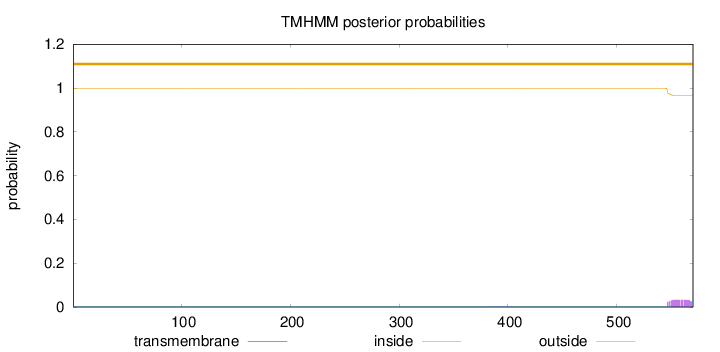
Topology
Length:
570
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.71508
Exp number, first 60 AAs:
0.00083
Total prob of N-in:
0.00077
outside
1 - 570
Population Genetic Test Statistics
Pi
269.916616
Theta
179.105441
Tajima's D
0.967684
CLR
0.312115
CSRT
0.653617319134043
Interpretation
Uncertain
