Gene
KWMTBOMO14123
Pre Gene Modal
BGIBMGA011193
Annotation
PREDICTED:_kinesin-like_protein_KIF23_[Plutella_xylostella]
Full name
Kinesin-like protein
Location in the cell
Nuclear Reliability : 3.165
Sequence
CDS
ATGAAGTCCGTTAAATCGAAACAGAGATTAAACGAGATGCGAACGCCGGCGCGTAGTAAACAAAATCTCGCTCAGGACCGCGATCCTGTACAAGTCTACTGTAGACTTCGCCCCATGCAAAGAGAAAGTGATGTGTCGTGTATGAAGATTATATCACCAACCACATTACTCCTAACGCCGCCAGCAACGGCGTTAAGCTACAGAAATGAAGGCAGCAAAACCATGAAATACACATTCAAAGAGATTTTCCCGCCGGAAGCGTCACAGCAAGAAGTTTTCGATAAAGTCTCACTGCCGTTAGTCGAAGGCTTGATTAAAGGTAAAAACGGCTTGTTATTCACCTACGGGGTCACGGGCAGCGGGAAAACGTTCACGATGACCGGCGAGCCCCAGAACTGCGGGATATTACCACGATGTTTGAATATTATTTTCAAAACTATTAACGATTTCCAAGCTCACAAATATGTGTTCAAACCGGACAAAATGAACATGTTCGATATTCAAAACGAAGCGGAGGCCATGCTAGAGAGGCAACAGGAACTTCACAAATTTAAAACTGGGAAGAAAAATAATTCGAATCCGGATTTAGCTATGGCATCCACGGATATAACTAAAGTAGAAGGTATCAATGAAGACAATCAGTACGCTGTGTTCGTTACATATGTAGAAATCTACAATAACAGTGTTTTTGACTTGTTAGAAGATGGTCCTCCGATTAGTAGGAACCTTCCAGTGAAGATAATAAGAGAAGATGCAGCCCATAATATGTATGTCCACGGGGTTACTGAAATCGAGATTAAGTCAGCTGAGGATGCCTTTGATGCTTTCTATTTAGGTTTGAAACGGAAACGAATGGCTCACACGACTCTGAACGCAGAATCAAGCCGAAGTCACTCAGTGTTCACAATACGATTAGTGCAGGCACCAGTTGACGAGATGGGAGAAGCTGTGATACAAAATAAAAAGTTCTTGAATATTAGTCAACTGAGTCTTGTTGATTTAGCTGGTAGCGAAAGAACTAACCGAACCAAAAACACAGGACAAAGACTAAGAGAAGCCGGGAATATTAATAAGTCATTGATGACTCTTAGAACCTGCTTAGAAGCGTTGAGAGAAAATCAAATTAACAGTGCTAATAACATGGTGCCCTATAGAGAGTCTAAAATTACTCATCTCTTCAAAAATTTCTTTGAAGGTGAAGGACAAGTAAGGATGATAGTCTGTGCAAATCCAAGAGCTGAAGATTATGATGAAACCCTCCAAGTGATGAGATTTGCTGAGATGGCAGCAGAAGTTGAAGTTGCTAAACCCCAAGTAATAGATGTCGCAACAACAATAGGTTTGATGTCTGGCCGCAGAAAGGCTAATAGACTCTTCACAACAGCTCGTGACAATATGGCGAGACCGGAGGCTAAAGATTTGGACTTAGATCTAGGTTTGGTATACAGCCTCGGGCCAGACTTCCCAAGCTTGGAACTGAATAGCTCTCAAGCTGCCCATATAATTAAGGAACTGATGGCACATTTAGAAATGAGAATCAGTCGCAGAGAAACACTTAAGAGAAGTAAGGCGGTGAAGGATGAAAGACTGAGGTCAGCCCTGTTGGATCTTGAAAAAGAATCATTAGAAGTGAGAAGTGAAAACTCATCATTGAGGGGACAGCTGGCAGCCTCTGAACGCAGGATTCGAGCTTTAGAAGAACGCTTGACAGCCACAGAAAATGCATCTCTCACGCACCAAAGAAAATTGGCAGAAATGACTGAATACACATCATCTTTAAAGAGAGATCTACAGGAAAAAGAATATTTGCTCAACCAGAATAAGTTTGAGAAAGAAAAGCAGAAAAAGATCCTTGAAAGAAAATACAACCACAAGATTGCCGCCGAACAGGAGAAGGCAAAAGAGATAAACACAGAAAAGGAAAGATTAAGAAATGCAGTTGAGGAAAAAGAAAACTGTTTACGTAAAGTCAAAGAAGCGTTAAATGTTGATATGGCAGACAGAGGAGACAACTTTGATTATCCACGCTCCACAACTGAAATTGGTGCTCAGACCAGTCATTTTACACCGGGCTCCACACCGAGAGCTCTACCCGCACACCTCTTGACCCCATTCAGTTCAGCTCCACAATCACGCGACACACCAACATCAAGAAGAGGGGTCGCTGTAGCGAATCCACGCCATAGACGGTCATTGTCAGCTGATGGTCGAGGCTGGGTGGAACACGCACCTAACAAGCTAGTCCCAATGGGCACAGTCATGAAGCCAAGCATGACGAATAGCAAAATCGTTAATAAACTGACAACGGTCAAAGACATTGCCAATTCGAAAACGTCAAAATACTGTCTTATATCGCAAGAGCAAGATACTGATGGTGAATTGGAGACGAAACTGTACAAAGGTGATGTTGTACCCACCTGCGGGGGAGGAGCTCAAGTTATATTCAATGATGTTGAAATGCTAAAACAGTTTTCTCCGACAAGGGAATCGCAGAGATTGTCCAGCAATAGCTGTGCTAGACATTCCGGCAAGAGGAGAGACACCGCTTCTTCACCACCACAAACACCATCAAACACCAGTAAGAAGCAGAGAGTTGATGACGAATGA
Protein
MKSVKSKQRLNEMRTPARSKQNLAQDRDPVQVYCRLRPMQRESDVSCMKIISPTTLLLTPPATALSYRNEGSKTMKYTFKEIFPPEASQQEVFDKVSLPLVEGLIKGKNGLLFTYGVTGSGKTFTMTGEPQNCGILPRCLNIIFKTINDFQAHKYVFKPDKMNMFDIQNEAEAMLERQQELHKFKTGKKNNSNPDLAMASTDITKVEGINEDNQYAVFVTYVEIYNNSVFDLLEDGPPISRNLPVKIIREDAAHNMYVHGVTEIEIKSAEDAFDAFYLGLKRKRMAHTTLNAESSRSHSVFTIRLVQAPVDEMGEAVIQNKKFLNISQLSLVDLAGSERTNRTKNTGQRLREAGNINKSLMTLRTCLEALRENQINSANNMVPYRESKITHLFKNFFEGEGQVRMIVCANPRAEDYDETLQVMRFAEMAAEVEVAKPQVIDVATTIGLMSGRRKANRLFTTARDNMARPEAKDLDLDLGLVYSLGPDFPSLELNSSQAAHIIKELMAHLEMRISRRETLKRSKAVKDERLRSALLDLEKESLEVRSENSSLRGQLAASERRIRALEERLTATENASLTHQRKLAEMTEYTSSLKRDLQEKEYLLNQNKFEKEKQKKILERKYNHKIAAEQEKAKEINTEKERLRNAVEEKENCLRKVKEALNVDMADRGDNFDYPRSTTEIGAQTSHFTPGSTPRALPAHLLTPFSSAPQSRDTPTSRRGVAVANPRHRRSLSADGRGWVEHAPNKLVPMGTVMKPSMTNSKIVNKLTTVKDIANSKTSKYCLISQEQDTDGELETKLYKGDVVPTCGGGAQVIFNDVEMLKQFSPTRESQRLSSNSCARHSGKRRDTASSPPQTPSNTSKKQRVDDE
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
H9JNT9
A0A2H1VKY3
A0A212F6J7
A0A194QRW4
A0A194QHQ3
A0A0L7L834
+ More
A0A2J7Q7W5 A0A1S4F2I9 Q17HE6 A0A336KG56 A0A088ARB0 A0A154PIV3 A0A0L7RC86 A0A182G352 A0A1Q3EZ45 A0A2A3EC26 K7J331 A0A0C9QSZ0 A0A0C9RES6 B0XEJ2 A0A1S4KBM6 A0A2P8XWW4 A0A1B0CIH0 A0A026VXP5 A0A1J1IYS6 A0A067QI98 A0A2J7R2U6 A0A1A9V461 A0A1A9YEF0 A0A1B0B7Y8 A0A182XZC2 A0A182M880 F4X1L2 B3M4X8 A0A158NKC9 A0A1B0FDS7 A0A182KS11 A0A182T8R3 A0A232FA23 A0A0M4EA03 A0A0L0CGW7 A0A182I3W1 A0A195BU49 B4HU29 A0A182RB17 A0A0J9RPA9 O62528 Q8SXT9 A0A182X6L2 Q9VZF5 A0A3B0J5C8 A0A182WIY8 B4PHS5 A0A182JTW2 Q7PT19 A0A182QC68 B3NC05 W5JJ93 B4L9B5 E2BRZ1 A0A1W4W694 B4LBP7 A0A1I8N1C8 A0A182PA07 B4J1G7 A0A034VAP7 A0A182NHC3 A0A151WKY7 A0A0K8VXY1 B4NLZ7 A0A182FG26 E2AZU9 A0A182IVR1 W8BXE8 E9JC91 A0A1I8Q1Q0 A0A195F658 A0A1I8Q1N0 A0A084VF61 Q29D63 B4H7X3 A0A1B6JW74 A0A2A3EH71 A0A088AM76 B4QQI5 A0A0L7RCZ6 B4IQZ0 A0A1B6F0N8 A0A154P1U0 A0A1B6D3A1 A0A067RLS7 A0A0A1WE31 A0A2J7Q7X6 E0VXW0 A0A0P5F4U9 A0A0P4XFN0 A0A0M8ZXY0 E9FXU2 A0A0N8DVU8 A0A0J7KVL1
A0A2J7Q7W5 A0A1S4F2I9 Q17HE6 A0A336KG56 A0A088ARB0 A0A154PIV3 A0A0L7RC86 A0A182G352 A0A1Q3EZ45 A0A2A3EC26 K7J331 A0A0C9QSZ0 A0A0C9RES6 B0XEJ2 A0A1S4KBM6 A0A2P8XWW4 A0A1B0CIH0 A0A026VXP5 A0A1J1IYS6 A0A067QI98 A0A2J7R2U6 A0A1A9V461 A0A1A9YEF0 A0A1B0B7Y8 A0A182XZC2 A0A182M880 F4X1L2 B3M4X8 A0A158NKC9 A0A1B0FDS7 A0A182KS11 A0A182T8R3 A0A232FA23 A0A0M4EA03 A0A0L0CGW7 A0A182I3W1 A0A195BU49 B4HU29 A0A182RB17 A0A0J9RPA9 O62528 Q8SXT9 A0A182X6L2 Q9VZF5 A0A3B0J5C8 A0A182WIY8 B4PHS5 A0A182JTW2 Q7PT19 A0A182QC68 B3NC05 W5JJ93 B4L9B5 E2BRZ1 A0A1W4W694 B4LBP7 A0A1I8N1C8 A0A182PA07 B4J1G7 A0A034VAP7 A0A182NHC3 A0A151WKY7 A0A0K8VXY1 B4NLZ7 A0A182FG26 E2AZU9 A0A182IVR1 W8BXE8 E9JC91 A0A1I8Q1Q0 A0A195F658 A0A1I8Q1N0 A0A084VF61 Q29D63 B4H7X3 A0A1B6JW74 A0A2A3EH71 A0A088AM76 B4QQI5 A0A0L7RCZ6 B4IQZ0 A0A1B6F0N8 A0A154P1U0 A0A1B6D3A1 A0A067RLS7 A0A0A1WE31 A0A2J7Q7X6 E0VXW0 A0A0P5F4U9 A0A0P4XFN0 A0A0M8ZXY0 E9FXU2 A0A0N8DVU8 A0A0J7KVL1
Pubmed
19121390
22118469
26354079
26227816
17510324
26483478
+ More
20075255 29403074 24508170 24845553 25244985 21719571 17994087 21347285 20966253 28648823 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 12364791 14747013 17210077 20920257 23761445 20798317 25315136 25348373 24495485 21282665 24438588 15632085 25830018 20566863 21292972
20075255 29403074 24508170 24845553 25244985 21719571 17994087 21347285 20966253 28648823 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 12364791 14747013 17210077 20920257 23761445 20798317 25315136 25348373 24495485 21282665 24438588 15632085 25830018 20566863 21292972
EMBL
BABH01009922
ODYU01003119
SOQ41477.1
AGBW02010004
OWR49361.1
KQ461198
+ More
KPJ06281.1 KQ458793 KPJ05073.1 JTDY01002331 KOB71648.1 NEVH01017000 PNF24674.1 CH477250 EAT46060.1 UFQS01000238 UFQT01000238 SSX01833.1 SSX22210.1 KQ434931 KZC11765.1 KQ414615 KOC68607.1 JXUM01140966 KQ569159 KXJ68749.1 GFDL01014473 JAV20572.1 KZ288289 PBC29287.1 GBYB01006844 JAG76611.1 GBYB01006845 JAG76612.1 DS232835 EDS26019.1 PYGN01001224 PSN36501.1 AJWK01013324 AJWK01013325 KK107796 EZA47599.1 CVRI01000063 CRL04318.1 KK853567 KDR06659.1 NEVH01007826 PNF35143.1 JXJN01009745 AXCM01001459 GL888531 EGI59670.1 CH902618 EDV40552.1 ADTU01002060 ADTU01002061 CCAG010004132 NNAY01000635 OXU27283.1 CP012525 ALC44016.1 JRES01000398 KNC31663.1 APCN01000225 KQ976403 KYM92134.1 CH480817 EDW50450.1 CM002912 KMY97542.1 AJ224882 CAA12181.1 AY084137 AAL89875.1 AE014296 BT053676 AAF47868.1 ACK77591.1 OUUW01000002 SPP76865.1 CM000159 EDW93384.1 AAAB01008807 EAA04675.5 AXCN02000896 CH954178 EDV50893.1 ADMH02001258 ETN63363.1 CH933816 EDW17290.1 GL450107 EFN81507.1 CH940647 EDW68674.2 CH916366 EDV95858.1 GAKP01019363 JAC39589.1 KQ982993 KYQ48514.1 GDHF01008571 JAI43743.1 CH964278 EDW85365.1 GL444277 EFN61078.1 GAMC01000540 JAC06016.1 GL771776 EFZ09558.1 KQ981805 KYN35549.1 ATLV01012362 KE524785 KFB36605.1 CH379070 EAL30551.2 CH479219 EDW34763.1 GECU01004235 JAT03472.1 KZ288248 PBC31125.1 CM000363 EDX09191.1 KOC68665.1 CH690061 EDW26657.1 GECZ01025999 JAS43770.1 KQ434796 KZC05792.1 GEDC01017172 JAS20126.1 KK852601 KDR20500.1 GBXI01017572 JAC96719.1 PNF24675.1 DS235841 EEB18216.1 GDIQ01265770 LRGB01000190 JAJ85954.1 KZS20441.1 GDIP01251516 JAI71885.1 KQ435802 KOX73197.1 GL732526 EFX88160.1 GDIQ01086884 JAN07853.1 LBMM01002800 KMQ94346.1
KPJ06281.1 KQ458793 KPJ05073.1 JTDY01002331 KOB71648.1 NEVH01017000 PNF24674.1 CH477250 EAT46060.1 UFQS01000238 UFQT01000238 SSX01833.1 SSX22210.1 KQ434931 KZC11765.1 KQ414615 KOC68607.1 JXUM01140966 KQ569159 KXJ68749.1 GFDL01014473 JAV20572.1 KZ288289 PBC29287.1 GBYB01006844 JAG76611.1 GBYB01006845 JAG76612.1 DS232835 EDS26019.1 PYGN01001224 PSN36501.1 AJWK01013324 AJWK01013325 KK107796 EZA47599.1 CVRI01000063 CRL04318.1 KK853567 KDR06659.1 NEVH01007826 PNF35143.1 JXJN01009745 AXCM01001459 GL888531 EGI59670.1 CH902618 EDV40552.1 ADTU01002060 ADTU01002061 CCAG010004132 NNAY01000635 OXU27283.1 CP012525 ALC44016.1 JRES01000398 KNC31663.1 APCN01000225 KQ976403 KYM92134.1 CH480817 EDW50450.1 CM002912 KMY97542.1 AJ224882 CAA12181.1 AY084137 AAL89875.1 AE014296 BT053676 AAF47868.1 ACK77591.1 OUUW01000002 SPP76865.1 CM000159 EDW93384.1 AAAB01008807 EAA04675.5 AXCN02000896 CH954178 EDV50893.1 ADMH02001258 ETN63363.1 CH933816 EDW17290.1 GL450107 EFN81507.1 CH940647 EDW68674.2 CH916366 EDV95858.1 GAKP01019363 JAC39589.1 KQ982993 KYQ48514.1 GDHF01008571 JAI43743.1 CH964278 EDW85365.1 GL444277 EFN61078.1 GAMC01000540 JAC06016.1 GL771776 EFZ09558.1 KQ981805 KYN35549.1 ATLV01012362 KE524785 KFB36605.1 CH379070 EAL30551.2 CH479219 EDW34763.1 GECU01004235 JAT03472.1 KZ288248 PBC31125.1 CM000363 EDX09191.1 KOC68665.1 CH690061 EDW26657.1 GECZ01025999 JAS43770.1 KQ434796 KZC05792.1 GEDC01017172 JAS20126.1 KK852601 KDR20500.1 GBXI01017572 JAC96719.1 PNF24675.1 DS235841 EEB18216.1 GDIQ01265770 LRGB01000190 JAJ85954.1 KZS20441.1 GDIP01251516 JAI71885.1 KQ435802 KOX73197.1 GL732526 EFX88160.1 GDIQ01086884 JAN07853.1 LBMM01002800 KMQ94346.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000037510
UP000235965
+ More
UP000008820 UP000005203 UP000076502 UP000053825 UP000069940 UP000249989 UP000242457 UP000002358 UP000002320 UP000245037 UP000092461 UP000053097 UP000183832 UP000027135 UP000078200 UP000092443 UP000092460 UP000076408 UP000075883 UP000007755 UP000007801 UP000005205 UP000092444 UP000075882 UP000075901 UP000215335 UP000092553 UP000037069 UP000075840 UP000078540 UP000001292 UP000075900 UP000076407 UP000000803 UP000268350 UP000075920 UP000002282 UP000075881 UP000007062 UP000075886 UP000008711 UP000000673 UP000009192 UP000008237 UP000192221 UP000008792 UP000095301 UP000075885 UP000001070 UP000075884 UP000075809 UP000007798 UP000069272 UP000000311 UP000075880 UP000095300 UP000078541 UP000030765 UP000001819 UP000008744 UP000000304 UP000009046 UP000076858 UP000053105 UP000000305 UP000036403
UP000008820 UP000005203 UP000076502 UP000053825 UP000069940 UP000249989 UP000242457 UP000002358 UP000002320 UP000245037 UP000092461 UP000053097 UP000183832 UP000027135 UP000078200 UP000092443 UP000092460 UP000076408 UP000075883 UP000007755 UP000007801 UP000005205 UP000092444 UP000075882 UP000075901 UP000215335 UP000092553 UP000037069 UP000075840 UP000078540 UP000001292 UP000075900 UP000076407 UP000000803 UP000268350 UP000075920 UP000002282 UP000075881 UP000007062 UP000075886 UP000008711 UP000000673 UP000009192 UP000008237 UP000192221 UP000008792 UP000095301 UP000075885 UP000001070 UP000075884 UP000075809 UP000007798 UP000069272 UP000000311 UP000075880 UP000095300 UP000078541 UP000030765 UP000001819 UP000008744 UP000000304 UP000009046 UP000076858 UP000053105 UP000000305 UP000036403
Pfam
Interpro
IPR038105
Kif23_Arf-bd_sf
+ More
IPR032384 Kif23_Arf-bd
IPR032924 KIF23
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR001752 Kinesin_motor_dom
IPR027640 Kinesin-like_fam
IPR036961 Kinesin_motor_dom_sf
IPR019176 Cytochrome_B561-rel
IPR009600 PIG-U
IPR036719 Neuro-gated_channel_TM_sf
IPR036734 Neur_chan_lig-bd_sf
IPR006202 Neur_chan_lig-bd
IPR006029 Neurotrans-gated_channel_TM
IPR018000 Neurotransmitter_ion_chnl_CS
IPR002130 Cyclophilin-type_PPIase_dom
IPR029000 Cyclophilin-like_dom_sf
IPR003938 K_chnl_volt-dep_EAG/ELK/ERG
IPR027359 Volt_channel_dom_sf
IPR030172 KCNH6
IPR003967 K_chnl_volt-dep_ERG
IPR000595 cNMP-bd_dom
IPR020892 Cyclophilin-type_PPIase_CS
IPR005821 Ion_trans_dom
IPR018490 cNMP-bd-like
IPR014710 RmlC-like_jellyroll
IPR016024 ARM-type_fold
IPR000014 PAS
IPR013122 PKD1_2_channel
IPR035965 PAS-like_dom_sf
IPR032384 Kif23_Arf-bd
IPR032924 KIF23
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR001752 Kinesin_motor_dom
IPR027640 Kinesin-like_fam
IPR036961 Kinesin_motor_dom_sf
IPR019176 Cytochrome_B561-rel
IPR009600 PIG-U
IPR036719 Neuro-gated_channel_TM_sf
IPR036734 Neur_chan_lig-bd_sf
IPR006202 Neur_chan_lig-bd
IPR006029 Neurotrans-gated_channel_TM
IPR018000 Neurotransmitter_ion_chnl_CS
IPR002130 Cyclophilin-type_PPIase_dom
IPR029000 Cyclophilin-like_dom_sf
IPR003938 K_chnl_volt-dep_EAG/ELK/ERG
IPR027359 Volt_channel_dom_sf
IPR030172 KCNH6
IPR003967 K_chnl_volt-dep_ERG
IPR000595 cNMP-bd_dom
IPR020892 Cyclophilin-type_PPIase_CS
IPR005821 Ion_trans_dom
IPR018490 cNMP-bd-like
IPR014710 RmlC-like_jellyroll
IPR016024 ARM-type_fold
IPR000014 PAS
IPR013122 PKD1_2_channel
IPR035965 PAS-like_dom_sf
SUPFAM
ProteinModelPortal
H9JNT9
A0A2H1VKY3
A0A212F6J7
A0A194QRW4
A0A194QHQ3
A0A0L7L834
+ More
A0A2J7Q7W5 A0A1S4F2I9 Q17HE6 A0A336KG56 A0A088ARB0 A0A154PIV3 A0A0L7RC86 A0A182G352 A0A1Q3EZ45 A0A2A3EC26 K7J331 A0A0C9QSZ0 A0A0C9RES6 B0XEJ2 A0A1S4KBM6 A0A2P8XWW4 A0A1B0CIH0 A0A026VXP5 A0A1J1IYS6 A0A067QI98 A0A2J7R2U6 A0A1A9V461 A0A1A9YEF0 A0A1B0B7Y8 A0A182XZC2 A0A182M880 F4X1L2 B3M4X8 A0A158NKC9 A0A1B0FDS7 A0A182KS11 A0A182T8R3 A0A232FA23 A0A0M4EA03 A0A0L0CGW7 A0A182I3W1 A0A195BU49 B4HU29 A0A182RB17 A0A0J9RPA9 O62528 Q8SXT9 A0A182X6L2 Q9VZF5 A0A3B0J5C8 A0A182WIY8 B4PHS5 A0A182JTW2 Q7PT19 A0A182QC68 B3NC05 W5JJ93 B4L9B5 E2BRZ1 A0A1W4W694 B4LBP7 A0A1I8N1C8 A0A182PA07 B4J1G7 A0A034VAP7 A0A182NHC3 A0A151WKY7 A0A0K8VXY1 B4NLZ7 A0A182FG26 E2AZU9 A0A182IVR1 W8BXE8 E9JC91 A0A1I8Q1Q0 A0A195F658 A0A1I8Q1N0 A0A084VF61 Q29D63 B4H7X3 A0A1B6JW74 A0A2A3EH71 A0A088AM76 B4QQI5 A0A0L7RCZ6 B4IQZ0 A0A1B6F0N8 A0A154P1U0 A0A1B6D3A1 A0A067RLS7 A0A0A1WE31 A0A2J7Q7X6 E0VXW0 A0A0P5F4U9 A0A0P4XFN0 A0A0M8ZXY0 E9FXU2 A0A0N8DVU8 A0A0J7KVL1
A0A2J7Q7W5 A0A1S4F2I9 Q17HE6 A0A336KG56 A0A088ARB0 A0A154PIV3 A0A0L7RC86 A0A182G352 A0A1Q3EZ45 A0A2A3EC26 K7J331 A0A0C9QSZ0 A0A0C9RES6 B0XEJ2 A0A1S4KBM6 A0A2P8XWW4 A0A1B0CIH0 A0A026VXP5 A0A1J1IYS6 A0A067QI98 A0A2J7R2U6 A0A1A9V461 A0A1A9YEF0 A0A1B0B7Y8 A0A182XZC2 A0A182M880 F4X1L2 B3M4X8 A0A158NKC9 A0A1B0FDS7 A0A182KS11 A0A182T8R3 A0A232FA23 A0A0M4EA03 A0A0L0CGW7 A0A182I3W1 A0A195BU49 B4HU29 A0A182RB17 A0A0J9RPA9 O62528 Q8SXT9 A0A182X6L2 Q9VZF5 A0A3B0J5C8 A0A182WIY8 B4PHS5 A0A182JTW2 Q7PT19 A0A182QC68 B3NC05 W5JJ93 B4L9B5 E2BRZ1 A0A1W4W694 B4LBP7 A0A1I8N1C8 A0A182PA07 B4J1G7 A0A034VAP7 A0A182NHC3 A0A151WKY7 A0A0K8VXY1 B4NLZ7 A0A182FG26 E2AZU9 A0A182IVR1 W8BXE8 E9JC91 A0A1I8Q1Q0 A0A195F658 A0A1I8Q1N0 A0A084VF61 Q29D63 B4H7X3 A0A1B6JW74 A0A2A3EH71 A0A088AM76 B4QQI5 A0A0L7RCZ6 B4IQZ0 A0A1B6F0N8 A0A154P1U0 A0A1B6D3A1 A0A067RLS7 A0A0A1WE31 A0A2J7Q7X6 E0VXW0 A0A0P5F4U9 A0A0P4XFN0 A0A0M8ZXY0 E9FXU2 A0A0N8DVU8 A0A0J7KVL1
PDB
5X3E
E-value=8.92176e-85,
Score=802
Ontologies
GO
GO:0072383
GO:0000915
GO:0005874
GO:0003777
GO:0005524
GO:0008017
GO:0005623
GO:0042765
GO:0016255
GO:0016021
GO:0005230
GO:0000281
GO:0070938
GO:1990023
GO:0005930
GO:0090090
GO:0001709
GO:0005828
GO:0032507
GO:0035324
GO:0030496
GO:0032154
GO:0007525
GO:0035323
GO:0000916
GO:1903562
GO:0070507
GO:0005634
GO:0098549
GO:0006457
GO:0005249
GO:0003755
GO:0034765
GO:0007018
GO:0005819
GO:0005871
GO:0007052
GO:0007422
GO:0016321
GO:0005813
GO:0051256
GO:0016887
GO:0019829
GO:0005507
GO:0004713
GO:0016491
GO:0015930
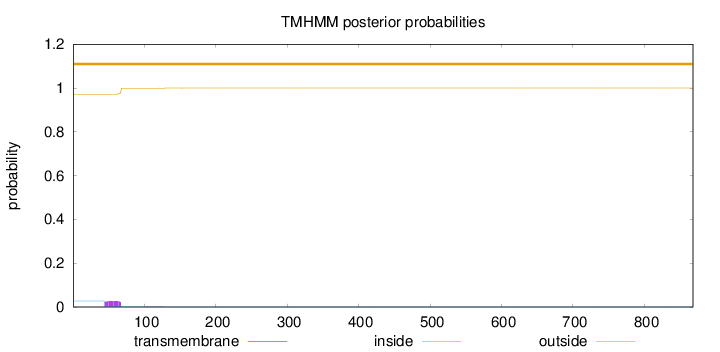
Topology
Length:
868
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.61444
Exp number, first 60 AAs:
0.42207
Total prob of N-in:
0.02785
outside
1 - 868
Population Genetic Test Statistics
Pi
314.259865
Theta
229.077536
Tajima's D
1.105895
CLR
0.345629
CSRT
0.690465476726164
Interpretation
Uncertain
