Gene
KWMTBOMO14122
Pre Gene Modal
BGIBMGA011393
Annotation
PREDICTED:_serine/threonine-protein_kinase_BRSK2_[Bombyx_mori]
Full name
DNA mismatch repair protein
Location in the cell
Cytoplasmic Reliability : 1.657 Mitochondrial Reliability : 1.411
Sequence
CDS
ATGGATAGTGGGTCGGCGACAACGACTGAGGCCTACCAATACGTAGGGCCCTTCCGGCTGGAGAAAACCTTAGGGAAGGGGCAAACAGGTCTAGTCAAGCTCGGGGTACATTGCGTGACGGGAAAGAAAGTGGCCATCAAGATTATCAACAGAGAGAAACTCAGCGAATCTGTGTTGACGAAGGTCGAAAGGGAAATAGCTATAATGAAATTAATTGAGCATCCCCACGTGCTCGGACTGTCCGACGTTTATGAAAATAAGAAATATTTGTATCTAGTTTTAGAGCACGTAAGCGGAGGCGAGCTGTTCGACTACTTAGTGAAGAAGGGCCGACTCACGCCGAAAGAGGCTCGGAGATTTTTCAGACAAATTATATCCGCCCTAGATTTCTGTCATAGTCACTCGATTTGCCACAGGGACTTAAAGCCGGAGAACTTGCTCCTAGACGAACGGAACAACATCAAGATTGCAGATTTCGGGATGGCCTCCCTCCAGCCGGCCGGGTCGCTGCTGGAAACTTCTTGCGGGAGTCCGCATTACGCTTGCCCCGAAGTCATACGGGTCATGCCCCAATATGTCCAGTTCCTTATAGATAATACATAA
Protein
MDSGSATTTEAYQYVGPFRLEKTLGKGQTGLVKLGVHCVTGKKVAIKIINREKLSESVLTKVEREIAIMKLIEHPHVLGLSDVYENKKYLYLVLEHVSGGELFDYLVKKGRLTPKEARRFFRQIISALDFCHSHSICHRDLKPENLLLDERNNIKIADFGMASLQPAGSLLETSCGSPHYACPEVIRVMPQYVQFLIDNT
Summary
Description
Component of the post-replicative DNA mismatch repair system (MMR).
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein-tyrosine phosphatase family. Non-receptor class dual specificity subfamily.
Belongs to the DNA mismatch repair MutS family.
Belongs to the protein-tyrosine phosphatase family. Non-receptor class dual specificity subfamily.
Belongs to the DNA mismatch repair MutS family.
Uniprot
A0A194RQQ3
A0A2W1BES3
A0A3S2NM17
B4PD12
A0A146L3K3
A0A1B0CQI3
+ More
A0A1A9W506 E1ZUV7 A0A088ARN5 A0A0L0CLZ6 A0A3L8E0G9 A0A182VAT9 A0A182J379 A0A151IGG1 A0A1S4EY07 A0A151WFE5 B4QLQ9 Q17M45 F4WM56 A0A195BDB0 A0A1I8Q331 A0A182KST8 A0A1I8Q339 A0A1I8N898 A0A195DCL6 A0A195FVZ0 Q7PV95 A0A182Y1X2 A0A158P3J7 A0A0L7RD24 A0A1B6D1M0 B4GRJ7 A0A084WB46 A0A2P2IGK7 A0A182MR70 K7IXS0 D2A4R1 A0A310SSE5 A0A0K8VF51 A0A0N8P0Y1 A0A0Q9X0L8 A0A3B0KMI4 A0A1W4VMZ6 A0A1W4VAZ2 A0A0R3P3T7 A0A3B0KA80 B5DPG7 M9PFM1 A0A0R1E7J2 A0A0Q5U654 A0A0R1ECS1 A0A0Q5UJT7 B3M9V3 A0A3B0KFC4 B4MXV5 M9PFS3 A0A1W4VBA3 Q9VUV4 A0A226CXU9 A0A1W4VPC2 B3NDN8 B4ITX3 A0A1B6D6G2 W8AXQ4 A0A1I8HJU7 W8BKG0 A0A0P5W749 A0A0P4XXJ2 E0VSU4 A0A0J9RVY5 A0A0P5IGB4 A0A2S2NLE4 B4J1V9 A0A0J9ULZ1 B4LDB8 E9FRJ6 A0A154PA70 A0A336N0U3 A0A2S2N893 A0A2H8TI99 A0A0A9WQZ3 A0A2R8QFR1 A0A182FUF5 A0A182HPY6 A0A182XDA3 R7UCJ1 A0A0P4XWS9 A0A0P5BTU3 A0A1A8UKI6 A0A0N7ZZ22 A0A182H6A7 A0A0P5ACD0 A0A0P4XEC5 A0A182RGL3 A0A0P6DKI6 A0A0P5BN49 A0A0P4Z1A7 A0A0P5ZPK0 A0A3B3BWE9 A0A0P5VP32
A0A1A9W506 E1ZUV7 A0A088ARN5 A0A0L0CLZ6 A0A3L8E0G9 A0A182VAT9 A0A182J379 A0A151IGG1 A0A1S4EY07 A0A151WFE5 B4QLQ9 Q17M45 F4WM56 A0A195BDB0 A0A1I8Q331 A0A182KST8 A0A1I8Q339 A0A1I8N898 A0A195DCL6 A0A195FVZ0 Q7PV95 A0A182Y1X2 A0A158P3J7 A0A0L7RD24 A0A1B6D1M0 B4GRJ7 A0A084WB46 A0A2P2IGK7 A0A182MR70 K7IXS0 D2A4R1 A0A310SSE5 A0A0K8VF51 A0A0N8P0Y1 A0A0Q9X0L8 A0A3B0KMI4 A0A1W4VMZ6 A0A1W4VAZ2 A0A0R3P3T7 A0A3B0KA80 B5DPG7 M9PFM1 A0A0R1E7J2 A0A0Q5U654 A0A0R1ECS1 A0A0Q5UJT7 B3M9V3 A0A3B0KFC4 B4MXV5 M9PFS3 A0A1W4VBA3 Q9VUV4 A0A226CXU9 A0A1W4VPC2 B3NDN8 B4ITX3 A0A1B6D6G2 W8AXQ4 A0A1I8HJU7 W8BKG0 A0A0P5W749 A0A0P4XXJ2 E0VSU4 A0A0J9RVY5 A0A0P5IGB4 A0A2S2NLE4 B4J1V9 A0A0J9ULZ1 B4LDB8 E9FRJ6 A0A154PA70 A0A336N0U3 A0A2S2N893 A0A2H8TI99 A0A0A9WQZ3 A0A2R8QFR1 A0A182FUF5 A0A182HPY6 A0A182XDA3 R7UCJ1 A0A0P4XWS9 A0A0P5BTU3 A0A1A8UKI6 A0A0N7ZZ22 A0A182H6A7 A0A0P5ACD0 A0A0P4XEC5 A0A182RGL3 A0A0P6DKI6 A0A0P5BN49 A0A0P4Z1A7 A0A0P5ZPK0 A0A3B3BWE9 A0A0P5VP32
Pubmed
26354079
28756777
17994087
17550304
26823975
20798317
+ More
26108605 30249741 17510324 21719571 20966253 25315136 12364791 25244985 21347285 24438588 20075255 18362917 19820115 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 26392545 20566863 22936249 21292972 25401762 23594743 23254933 26483478 29451363
26108605 30249741 17510324 21719571 20966253 25315136 12364791 25244985 21347285 24438588 20075255 18362917 19820115 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 26392545 20566863 22936249 21292972 25401762 23594743 23254933 26483478 29451363
EMBL
KQ459875
KPJ19640.1
KZ150153
PZC72811.1
RSAL01000005
RVE54391.1
+ More
CM000159 EDW94944.2 GDHC01015535 JAQ03094.1 AJWK01023619 GL434297 EFN75049.1 JRES01000306 KNC32459.1 QOIP01000002 RLU25983.1 KQ977726 KYN00311.1 KQ983219 KYQ46552.1 CM000363 EDX10608.1 CH477208 EAT47800.1 GL888217 EGI64700.1 KQ976519 KYM82175.1 KQ980989 KYN10582.1 KQ981264 KYN44009.1 AAAB01008986 EAA00228.5 ADTU01001329 KQ414614 KOC68862.1 GEDC01017724 JAS19574.1 CH479188 EDW40382.1 ATLV01022312 KE525331 KFB47440.1 IACF01007493 LAB73076.1 AXCM01000926 KQ971344 EFA05190.2 KQ760869 OAD58767.1 GDHF01014852 JAI37462.1 CH902618 KPU78388.1 CH963876 KRF98534.1 OUUW01000009 SPP85018.1 CH379069 KRT07785.1 SPP85020.1 EDY73513.1 KRT07784.1 AE014296 AGB94595.1 CH891734 KRK05248.1 CH954178 KQS44153.1 KRK05249.1 KQS44154.1 EDV40144.1 SPP85019.1 EDW76874.1 AGB94594.1 AAF49569.3 LNIX01000048 OXA38155.1 EDV52171.1 EDW99836.1 KRK05250.1 GEDC01016026 JAS21272.1 GAMC01012795 JAB93760.1 GAMC01012794 JAB93761.1 GDIP01090371 JAM13344.1 GDIP01235372 JAI88029.1 DS235758 EEB16450.1 CM002912 KMY99876.1 GDIQ01212828 JAK38897.1 GGMR01005329 MBY17948.1 CH916366 EDV98039.1 KMY99875.1 CH940647 EDW71025.1 GL732523 EFX90176.1 KQ434856 KZC08733.1 UFQS01004145 UFQT01000829 UFQT01004145 SSX16209.1 SSX27442.1 SSX35535.1 GGMR01000824 MBY13443.1 GFXV01000996 GFXV01001317 GFXV01001936 MBW12801.1 MBW13122.1 MBW13741.1 GBHO01033758 JAG09846.1 AL935197 APCN01003284 AMQN01001733 KB305005 ELU01498.1 GDIP01239675 JAI83726.1 GDIP01180711 JAJ42691.1 HAEJ01007375 SBS47832.1 GDIP01198561 JAJ24841.1 JXUM01026850 JXUM01026851 JXUM01026852 JXUM01026853 JXUM01026854 JXUM01026855 JXUM01026856 JXUM01026857 JXUM01026858 JXUM01026859 KQ560769 KXJ80957.1 GDIP01201168 JAJ22234.1 GDIP01242306 JAI81095.1 GDIP01169411 GDIQ01076381 JAJ53991.1 JAN18356.1 GDIP01182362 JAJ41040.1 GDIP01219814 JAJ03588.1 GDIP01054080 JAM49635.1 GDIP01097420 JAM06295.1
CM000159 EDW94944.2 GDHC01015535 JAQ03094.1 AJWK01023619 GL434297 EFN75049.1 JRES01000306 KNC32459.1 QOIP01000002 RLU25983.1 KQ977726 KYN00311.1 KQ983219 KYQ46552.1 CM000363 EDX10608.1 CH477208 EAT47800.1 GL888217 EGI64700.1 KQ976519 KYM82175.1 KQ980989 KYN10582.1 KQ981264 KYN44009.1 AAAB01008986 EAA00228.5 ADTU01001329 KQ414614 KOC68862.1 GEDC01017724 JAS19574.1 CH479188 EDW40382.1 ATLV01022312 KE525331 KFB47440.1 IACF01007493 LAB73076.1 AXCM01000926 KQ971344 EFA05190.2 KQ760869 OAD58767.1 GDHF01014852 JAI37462.1 CH902618 KPU78388.1 CH963876 KRF98534.1 OUUW01000009 SPP85018.1 CH379069 KRT07785.1 SPP85020.1 EDY73513.1 KRT07784.1 AE014296 AGB94595.1 CH891734 KRK05248.1 CH954178 KQS44153.1 KRK05249.1 KQS44154.1 EDV40144.1 SPP85019.1 EDW76874.1 AGB94594.1 AAF49569.3 LNIX01000048 OXA38155.1 EDV52171.1 EDW99836.1 KRK05250.1 GEDC01016026 JAS21272.1 GAMC01012795 JAB93760.1 GAMC01012794 JAB93761.1 GDIP01090371 JAM13344.1 GDIP01235372 JAI88029.1 DS235758 EEB16450.1 CM002912 KMY99876.1 GDIQ01212828 JAK38897.1 GGMR01005329 MBY17948.1 CH916366 EDV98039.1 KMY99875.1 CH940647 EDW71025.1 GL732523 EFX90176.1 KQ434856 KZC08733.1 UFQS01004145 UFQT01000829 UFQT01004145 SSX16209.1 SSX27442.1 SSX35535.1 GGMR01000824 MBY13443.1 GFXV01000996 GFXV01001317 GFXV01001936 MBW12801.1 MBW13122.1 MBW13741.1 GBHO01033758 JAG09846.1 AL935197 APCN01003284 AMQN01001733 KB305005 ELU01498.1 GDIP01239675 JAI83726.1 GDIP01180711 JAJ42691.1 HAEJ01007375 SBS47832.1 GDIP01198561 JAJ24841.1 JXUM01026850 JXUM01026851 JXUM01026852 JXUM01026853 JXUM01026854 JXUM01026855 JXUM01026856 JXUM01026857 JXUM01026858 JXUM01026859 KQ560769 KXJ80957.1 GDIP01201168 JAJ22234.1 GDIP01242306 JAI81095.1 GDIP01169411 GDIQ01076381 JAJ53991.1 JAN18356.1 GDIP01182362 JAJ41040.1 GDIP01219814 JAJ03588.1 GDIP01054080 JAM49635.1 GDIP01097420 JAM06295.1
Proteomes
UP000053240
UP000283053
UP000002282
UP000092461
UP000091820
UP000000311
+ More
UP000005203 UP000037069 UP000279307 UP000075903 UP000075880 UP000078542 UP000075809 UP000000304 UP000008820 UP000007755 UP000078540 UP000095300 UP000075882 UP000095301 UP000078492 UP000078541 UP000007062 UP000076408 UP000005205 UP000053825 UP000008744 UP000030765 UP000075883 UP000002358 UP000007266 UP000007801 UP000007798 UP000268350 UP000192221 UP000001819 UP000000803 UP000008711 UP000198287 UP000095280 UP000009046 UP000001070 UP000008792 UP000000305 UP000076502 UP000000437 UP000069272 UP000075840 UP000076407 UP000014760 UP000069940 UP000249989 UP000075900 UP000261560
UP000005203 UP000037069 UP000279307 UP000075903 UP000075880 UP000078542 UP000075809 UP000000304 UP000008820 UP000007755 UP000078540 UP000095300 UP000075882 UP000095301 UP000078492 UP000078541 UP000007062 UP000076408 UP000005205 UP000053825 UP000008744 UP000030765 UP000075883 UP000002358 UP000007266 UP000007801 UP000007798 UP000268350 UP000192221 UP000001819 UP000000803 UP000008711 UP000198287 UP000095280 UP000009046 UP000001070 UP000008792 UP000000305 UP000076502 UP000000437 UP000069272 UP000075840 UP000076407 UP000014760 UP000069940 UP000249989 UP000075900 UP000261560
Pfam
Interpro
IPR017441
Protein_kinase_ATP_BS
+ More
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR017348 PIM1/2/3
IPR015940 UBA
IPR006020 PTB/PI_dom
IPR011993 PH-like_dom_sf
IPR001478 PDZ
IPR036034 PDZ_sf
IPR000387 TYR_PHOSPHATASE_dom
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000340 Dual-sp_phosphatase_cat-dom
IPR029021 Prot-tyrosine_phosphatase-like
IPR020417 Atypical_DUSP
IPR017261 DNA_mismatch_repair_MutS/MSH
IPR036187 DNA_mismatch_repair_MutS_sf
IPR007861 DNA_mismatch_repair_MutS_clamp
IPR036678 MutS_con_dom_sf
IPR000432 DNA_mismatch_repair_MutS_C
IPR007696 DNA_mismatch_repair_MutS_core
IPR007860 DNA_mmatch_repair_MutS_con_dom
IPR027417 P-loop_NTPase
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR017348 PIM1/2/3
IPR015940 UBA
IPR006020 PTB/PI_dom
IPR011993 PH-like_dom_sf
IPR001478 PDZ
IPR036034 PDZ_sf
IPR000387 TYR_PHOSPHATASE_dom
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000340 Dual-sp_phosphatase_cat-dom
IPR029021 Prot-tyrosine_phosphatase-like
IPR020417 Atypical_DUSP
IPR017261 DNA_mismatch_repair_MutS/MSH
IPR036187 DNA_mismatch_repair_MutS_sf
IPR007861 DNA_mismatch_repair_MutS_clamp
IPR036678 MutS_con_dom_sf
IPR000432 DNA_mismatch_repair_MutS_C
IPR007696 DNA_mismatch_repair_MutS_core
IPR007860 DNA_mmatch_repair_MutS_con_dom
IPR027417 P-loop_NTPase
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A194RQQ3
A0A2W1BES3
A0A3S2NM17
B4PD12
A0A146L3K3
A0A1B0CQI3
+ More
A0A1A9W506 E1ZUV7 A0A088ARN5 A0A0L0CLZ6 A0A3L8E0G9 A0A182VAT9 A0A182J379 A0A151IGG1 A0A1S4EY07 A0A151WFE5 B4QLQ9 Q17M45 F4WM56 A0A195BDB0 A0A1I8Q331 A0A182KST8 A0A1I8Q339 A0A1I8N898 A0A195DCL6 A0A195FVZ0 Q7PV95 A0A182Y1X2 A0A158P3J7 A0A0L7RD24 A0A1B6D1M0 B4GRJ7 A0A084WB46 A0A2P2IGK7 A0A182MR70 K7IXS0 D2A4R1 A0A310SSE5 A0A0K8VF51 A0A0N8P0Y1 A0A0Q9X0L8 A0A3B0KMI4 A0A1W4VMZ6 A0A1W4VAZ2 A0A0R3P3T7 A0A3B0KA80 B5DPG7 M9PFM1 A0A0R1E7J2 A0A0Q5U654 A0A0R1ECS1 A0A0Q5UJT7 B3M9V3 A0A3B0KFC4 B4MXV5 M9PFS3 A0A1W4VBA3 Q9VUV4 A0A226CXU9 A0A1W4VPC2 B3NDN8 B4ITX3 A0A1B6D6G2 W8AXQ4 A0A1I8HJU7 W8BKG0 A0A0P5W749 A0A0P4XXJ2 E0VSU4 A0A0J9RVY5 A0A0P5IGB4 A0A2S2NLE4 B4J1V9 A0A0J9ULZ1 B4LDB8 E9FRJ6 A0A154PA70 A0A336N0U3 A0A2S2N893 A0A2H8TI99 A0A0A9WQZ3 A0A2R8QFR1 A0A182FUF5 A0A182HPY6 A0A182XDA3 R7UCJ1 A0A0P4XWS9 A0A0P5BTU3 A0A1A8UKI6 A0A0N7ZZ22 A0A182H6A7 A0A0P5ACD0 A0A0P4XEC5 A0A182RGL3 A0A0P6DKI6 A0A0P5BN49 A0A0P4Z1A7 A0A0P5ZPK0 A0A3B3BWE9 A0A0P5VP32
A0A1A9W506 E1ZUV7 A0A088ARN5 A0A0L0CLZ6 A0A3L8E0G9 A0A182VAT9 A0A182J379 A0A151IGG1 A0A1S4EY07 A0A151WFE5 B4QLQ9 Q17M45 F4WM56 A0A195BDB0 A0A1I8Q331 A0A182KST8 A0A1I8Q339 A0A1I8N898 A0A195DCL6 A0A195FVZ0 Q7PV95 A0A182Y1X2 A0A158P3J7 A0A0L7RD24 A0A1B6D1M0 B4GRJ7 A0A084WB46 A0A2P2IGK7 A0A182MR70 K7IXS0 D2A4R1 A0A310SSE5 A0A0K8VF51 A0A0N8P0Y1 A0A0Q9X0L8 A0A3B0KMI4 A0A1W4VMZ6 A0A1W4VAZ2 A0A0R3P3T7 A0A3B0KA80 B5DPG7 M9PFM1 A0A0R1E7J2 A0A0Q5U654 A0A0R1ECS1 A0A0Q5UJT7 B3M9V3 A0A3B0KFC4 B4MXV5 M9PFS3 A0A1W4VBA3 Q9VUV4 A0A226CXU9 A0A1W4VPC2 B3NDN8 B4ITX3 A0A1B6D6G2 W8AXQ4 A0A1I8HJU7 W8BKG0 A0A0P5W749 A0A0P4XXJ2 E0VSU4 A0A0J9RVY5 A0A0P5IGB4 A0A2S2NLE4 B4J1V9 A0A0J9ULZ1 B4LDB8 E9FRJ6 A0A154PA70 A0A336N0U3 A0A2S2N893 A0A2H8TI99 A0A0A9WQZ3 A0A2R8QFR1 A0A182FUF5 A0A182HPY6 A0A182XDA3 R7UCJ1 A0A0P4XWS9 A0A0P5BTU3 A0A1A8UKI6 A0A0N7ZZ22 A0A182H6A7 A0A0P5ACD0 A0A0P4XEC5 A0A182RGL3 A0A0P6DKI6 A0A0P5BN49 A0A0P4Z1A7 A0A0P5ZPK0 A0A3B3BWE9 A0A0P5VP32
PDB
4YOM
E-value=3.10117e-92,
Score=859
Ontologies
GO
Topology
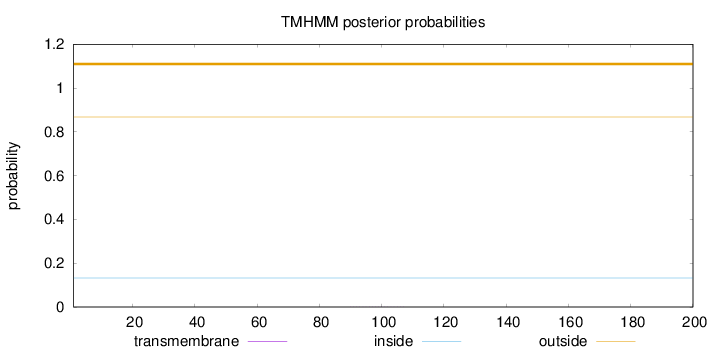
Length:
200
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00550999999999999
Exp number, first 60 AAs:
0.00179
Total prob of N-in:
0.13265
outside
1 - 200
Population Genetic Test Statistics
Pi
350.221568
Theta
172.840822
Tajima's D
4.019002
CLR
0.340368
CSRT
0.998650067496625
Interpretation
Uncertain
