Pre Gene Modal
BGIBMGA011196
Annotation
uncharacterized_protein_LOC692883_[Bombyx_mori]
Full name
Eukaryotic translation initiation factor 3 subunit H
Location in the cell
Extracellular Reliability : 1.567
Sequence
CDS
ATGTGGCCATTAATACTGCAGTTCATGAGAGCTAATGCGGCTTACATAACCCTCCCGGTTGCGGCTTTAGTAGGAGTCATAGGATACAACTTGGAAAACATATTGTCCGATAAATACACACCGTATAACAAGTCCATCGAAGACCAACGTGTAGACAGGCTAACGGATGAAGTATTGAAAGACCCAACCAATGTGAAGAAGCTGAAATATCAGGAGAATGTGCTCGGAAAGAATGTTTCCCCTTCGCTAGAAAAAGACTAG
Protein
MWPLILQFMRANAAYITLPVAALVGVIGYNLENILSDKYTPYNKSIEDQRVDRLTDEVLKDPTNVKKLKYQENVLGKNVSPSLEKD
Summary
Description
Component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is involved in protein synthesis of a specialized repertoire of mRNAs and, together with other initiation factors, stimulates binding of mRNA and methionyl-tRNAi to the 40S ribosome. The eIF-3 complex specifically targets and initiates translation of a subset of mRNAs involved in cell proliferation.
Subunit
Component of the eukaryotic translation initiation factor 3 (eIF-3) complex.
Similarity
Belongs to the eIF-3 subunit H family.
Uniprot
Q2F5Z6
A0A212FKD6
S4PCW0
A0A194RQG7
A0A3S2P159
A0A2H1V4P5
+ More
T1E8A3 A0A2W1BCV6 A0A2M4AYZ8 A0A023ECA3 A0A2M3ZDC0 A0A2M4C5U6 W5JDH7 A0A182FDQ6 Q16HH3 A0A023EDV2 A0A182I3Z5 A0A182V056 A0A182X8W3 A0A182LL31 A0NES5 A0A182YP71 A0A182PFM7 T1E3E8 A0A182NID6 A0A182WC76 A0A182KJ00 A0A1Q3FDD5 A0A182RKG9 B0WMQ2 A0A1B0DMB2 A0A336K369 U5EXY5 A0A1L8DAJ8 A0A1L8EIU9 A0A139WGT4 A0A1B0BKR2 A0A1L8EIZ0 A0A1I8PEU0 A0A1L8EJ21 A0A0L0C1G1 A0A1B0FMX2 A0A194PYL6 A0A1I8NAA4 A0A3B0J1D0 B4LPX7 A0A1W4UZH9 A0A0M4EFQ9 B4J911 B3MG55 A0A2J7QJK2 B4P626 B4HNJ6 B4QBM9 Q6IGN6 B4KR07 B3NSE3 B5E026 B4GAP4 A0A067QWV8 B4MYA6 A0A2P8Y945 A0A0P6H9R9 A0A224XRU3 A0A1D2MKC7 A0A0N7Z9A7 R4G494 A0A0P6BQH1 A0A069DMH8 A6YPT8 E9GMA9 A0A2R7WHZ2 A0A170V1M1 A0A151I8Y4 A0A151WRH4 A0A195B060 A0A158NKU1 A0A226DAL8 A0A151JBG9 A0A195FCK5 T1JGN7 A0A232EIQ7 A0A2L2YAM7 A0A0K8RPH8 E2AC70 A0A023FMX9 B7PSB5 A0A131XEY7 V4B537 A0A2C9JYU2 A0A023G6I7 A0A131YFU9 F0JAE0 L7M5L1 A0A1E1XTR7 A0A0L8GF32 A0A026X231
T1E8A3 A0A2W1BCV6 A0A2M4AYZ8 A0A023ECA3 A0A2M3ZDC0 A0A2M4C5U6 W5JDH7 A0A182FDQ6 Q16HH3 A0A023EDV2 A0A182I3Z5 A0A182V056 A0A182X8W3 A0A182LL31 A0NES5 A0A182YP71 A0A182PFM7 T1E3E8 A0A182NID6 A0A182WC76 A0A182KJ00 A0A1Q3FDD5 A0A182RKG9 B0WMQ2 A0A1B0DMB2 A0A336K369 U5EXY5 A0A1L8DAJ8 A0A1L8EIU9 A0A139WGT4 A0A1B0BKR2 A0A1L8EIZ0 A0A1I8PEU0 A0A1L8EJ21 A0A0L0C1G1 A0A1B0FMX2 A0A194PYL6 A0A1I8NAA4 A0A3B0J1D0 B4LPX7 A0A1W4UZH9 A0A0M4EFQ9 B4J911 B3MG55 A0A2J7QJK2 B4P626 B4HNJ6 B4QBM9 Q6IGN6 B4KR07 B3NSE3 B5E026 B4GAP4 A0A067QWV8 B4MYA6 A0A2P8Y945 A0A0P6H9R9 A0A224XRU3 A0A1D2MKC7 A0A0N7Z9A7 R4G494 A0A0P6BQH1 A0A069DMH8 A6YPT8 E9GMA9 A0A2R7WHZ2 A0A170V1M1 A0A151I8Y4 A0A151WRH4 A0A195B060 A0A158NKU1 A0A226DAL8 A0A151JBG9 A0A195FCK5 T1JGN7 A0A232EIQ7 A0A2L2YAM7 A0A0K8RPH8 E2AC70 A0A023FMX9 B7PSB5 A0A131XEY7 V4B537 A0A2C9JYU2 A0A023G6I7 A0A131YFU9 F0JAE0 L7M5L1 A0A1E1XTR7 A0A0L8GF32 A0A026X231
Pubmed
19121390
22118469
23622113
26354079
28756777
24945155
+ More
26483478 20920257 23761445 17510324 20966253 12364791 14747013 17210077 25244985 24330624 18362917 19820115 26108605 25315136 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 14709175 16110336 17569856 17569867 26109357 26109356 15632085 24845553 29403074 27289101 27129103 26334808 18207082 21292972 21347285 28648823 26561354 20798317 28049606 23254933 15562597 26830274 21362191 25576852 29209593 24508170 30249741
26483478 20920257 23761445 17510324 20966253 12364791 14747013 17210077 25244985 24330624 18362917 19820115 26108605 25315136 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 14709175 16110336 17569856 17569867 26109357 26109356 15632085 24845553 29403074 27289101 27129103 26334808 18207082 21292972 21347285 28648823 26561354 20798317 28049606 23254933 15562597 26830274 21362191 25576852 29209593 24508170 30249741
EMBL
BABH01009914
DQ311276
ABD36221.1
AGBW02008068
OWR54198.1
GAIX01003936
+ More
JAA88624.1 KQ459875 KPJ19644.1 RSAL01000005 RVE54393.1 ODYU01000658 SOQ35813.1 GAMD01002352 JAA99238.1 KZ150153 PZC72808.1 GGFK01012621 MBW45942.1 JXUM01053738 GAPW01006646 GAPW01006645 KQ561789 JAC06952.1 KXJ77529.1 GGFM01005806 MBW26557.1 GGFJ01011197 MBW60338.1 ADMH02001842 ETN60935.1 CH478170 EAT33695.1 GAPW01006647 JAC06951.1 APCN01002254 AAAB01008960 EAU76596.1 GALA01000420 JAA94432.1 GFDL01009478 JAV25567.1 DS232000 EDS31137.1 AJVK01074833 UFQS01000031 UFQT01000031 SSW97855.1 SSX18241.1 GANO01002293 JAB57578.1 GFDF01010689 JAV03395.1 GFDG01000172 JAV18627.1 KQ971345 KYB27065.1 JXJN01015994 GFDG01000171 JAV18628.1 GFDG01000173 JAV18626.1 JRES01001012 KNC26125.1 CCAG010018035 KQ459593 KPI96245.1 OUUW01000001 SPP74697.1 CH940648 EDW61317.2 CP012524 ALC41097.1 CH916367 EDW01360.1 CH902619 EDV36750.1 NEVH01013555 PNF28749.1 CM000158 EDW90901.2 CH480816 EDW47432.1 CM000362 CM002911 EDX06646.1 KMY93018.1 BT029595 BT029612 BT030156 BT030193 AE013599 BK003730 ABL75654.1 ABL75671.1 ABN49295.1 ABN49332.1 ABV53767.1 DAA02428.1 CH933808 EDW10363.1 CH954179 EDV56445.2 CM000071 EDY68910.2 CH479181 EDW31996.1 KK852869 KDR14664.1 CH963894 EDW77095.1 PYGN01000790 PSN40785.1 GDIQ01022443 JAN72294.1 GFTR01001219 JAW15207.1 LJIJ01001033 ODM93274.1 GDKW01001075 JAI55520.1 ACPB03003261 GAHY01000961 JAA76549.1 GDIP01011256 JAM92459.1 GBGD01003636 JAC85253.1 EF639099 ABR27984.1 GL732552 EFX79422.1 KK854856 PTY19263.1 GEMB01007096 JAR96272.1 KQ978323 KYM95113.1 KQ982805 KYQ50466.1 KQ976692 KYM77863.1 ADTU01018855 LNIX01000025 OXA42595.1 KQ979182 KYN22339.1 KQ981693 KYN37774.1 JH432211 NNAY01004173 OXU18243.1 IAAA01015390 LAA05206.1 GADI01000838 JAA72970.1 GL438428 EFN68964.1 GBBK01001375 JAC23107.1 ABJB010194957 DS777490 EEC09487.1 GEFH01002497 JAP66084.1 KB203660 ESO83574.1 GBBM01005986 GBBM01005985 GBBM01005984 JAC29434.1 GEDV01011756 JAP76801.1 BK007841 DAA34779.1 GACK01006586 JAA58448.1 GFAA01000740 JAU02695.1 KQ422303 KOF75140.1 KK107021 QOIP01000011 EZA62347.1 RLU17098.1
JAA88624.1 KQ459875 KPJ19644.1 RSAL01000005 RVE54393.1 ODYU01000658 SOQ35813.1 GAMD01002352 JAA99238.1 KZ150153 PZC72808.1 GGFK01012621 MBW45942.1 JXUM01053738 GAPW01006646 GAPW01006645 KQ561789 JAC06952.1 KXJ77529.1 GGFM01005806 MBW26557.1 GGFJ01011197 MBW60338.1 ADMH02001842 ETN60935.1 CH478170 EAT33695.1 GAPW01006647 JAC06951.1 APCN01002254 AAAB01008960 EAU76596.1 GALA01000420 JAA94432.1 GFDL01009478 JAV25567.1 DS232000 EDS31137.1 AJVK01074833 UFQS01000031 UFQT01000031 SSW97855.1 SSX18241.1 GANO01002293 JAB57578.1 GFDF01010689 JAV03395.1 GFDG01000172 JAV18627.1 KQ971345 KYB27065.1 JXJN01015994 GFDG01000171 JAV18628.1 GFDG01000173 JAV18626.1 JRES01001012 KNC26125.1 CCAG010018035 KQ459593 KPI96245.1 OUUW01000001 SPP74697.1 CH940648 EDW61317.2 CP012524 ALC41097.1 CH916367 EDW01360.1 CH902619 EDV36750.1 NEVH01013555 PNF28749.1 CM000158 EDW90901.2 CH480816 EDW47432.1 CM000362 CM002911 EDX06646.1 KMY93018.1 BT029595 BT029612 BT030156 BT030193 AE013599 BK003730 ABL75654.1 ABL75671.1 ABN49295.1 ABN49332.1 ABV53767.1 DAA02428.1 CH933808 EDW10363.1 CH954179 EDV56445.2 CM000071 EDY68910.2 CH479181 EDW31996.1 KK852869 KDR14664.1 CH963894 EDW77095.1 PYGN01000790 PSN40785.1 GDIQ01022443 JAN72294.1 GFTR01001219 JAW15207.1 LJIJ01001033 ODM93274.1 GDKW01001075 JAI55520.1 ACPB03003261 GAHY01000961 JAA76549.1 GDIP01011256 JAM92459.1 GBGD01003636 JAC85253.1 EF639099 ABR27984.1 GL732552 EFX79422.1 KK854856 PTY19263.1 GEMB01007096 JAR96272.1 KQ978323 KYM95113.1 KQ982805 KYQ50466.1 KQ976692 KYM77863.1 ADTU01018855 LNIX01000025 OXA42595.1 KQ979182 KYN22339.1 KQ981693 KYN37774.1 JH432211 NNAY01004173 OXU18243.1 IAAA01015390 LAA05206.1 GADI01000838 JAA72970.1 GL438428 EFN68964.1 GBBK01001375 JAC23107.1 ABJB010194957 DS777490 EEC09487.1 GEFH01002497 JAP66084.1 KB203660 ESO83574.1 GBBM01005986 GBBM01005985 GBBM01005984 JAC29434.1 GEDV01011756 JAP76801.1 BK007841 DAA34779.1 GACK01006586 JAA58448.1 GFAA01000740 JAU02695.1 KQ422303 KOF75140.1 KK107021 QOIP01000011 EZA62347.1 RLU17098.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000283053
UP000069940
UP000249989
+ More
UP000000673 UP000069272 UP000008820 UP000075840 UP000075903 UP000076407 UP000075882 UP000007062 UP000076408 UP000075885 UP000075884 UP000075920 UP000075881 UP000075900 UP000002320 UP000092462 UP000007266 UP000092460 UP000095300 UP000037069 UP000092444 UP000053268 UP000095301 UP000268350 UP000008792 UP000192221 UP000092553 UP000001070 UP000007801 UP000235965 UP000002282 UP000001292 UP000000304 UP000000803 UP000009192 UP000008711 UP000001819 UP000008744 UP000027135 UP000007798 UP000245037 UP000094527 UP000015103 UP000000305 UP000078542 UP000075809 UP000078540 UP000005205 UP000198287 UP000078492 UP000078541 UP000215335 UP000000311 UP000001555 UP000030746 UP000076420 UP000053454 UP000053097 UP000279307
UP000000673 UP000069272 UP000008820 UP000075840 UP000075903 UP000076407 UP000075882 UP000007062 UP000076408 UP000075885 UP000075884 UP000075920 UP000075881 UP000075900 UP000002320 UP000092462 UP000007266 UP000092460 UP000095300 UP000037069 UP000092444 UP000053268 UP000095301 UP000268350 UP000008792 UP000192221 UP000092553 UP000001070 UP000007801 UP000235965 UP000002282 UP000001292 UP000000304 UP000000803 UP000009192 UP000008711 UP000001819 UP000008744 UP000027135 UP000007798 UP000245037 UP000094527 UP000015103 UP000000305 UP000078542 UP000075809 UP000078540 UP000005205 UP000198287 UP000078492 UP000078541 UP000215335 UP000000311 UP000001555 UP000030746 UP000076420 UP000053454 UP000053097 UP000279307
ProteinModelPortal
Q2F5Z6
A0A212FKD6
S4PCW0
A0A194RQG7
A0A3S2P159
A0A2H1V4P5
+ More
T1E8A3 A0A2W1BCV6 A0A2M4AYZ8 A0A023ECA3 A0A2M3ZDC0 A0A2M4C5U6 W5JDH7 A0A182FDQ6 Q16HH3 A0A023EDV2 A0A182I3Z5 A0A182V056 A0A182X8W3 A0A182LL31 A0NES5 A0A182YP71 A0A182PFM7 T1E3E8 A0A182NID6 A0A182WC76 A0A182KJ00 A0A1Q3FDD5 A0A182RKG9 B0WMQ2 A0A1B0DMB2 A0A336K369 U5EXY5 A0A1L8DAJ8 A0A1L8EIU9 A0A139WGT4 A0A1B0BKR2 A0A1L8EIZ0 A0A1I8PEU0 A0A1L8EJ21 A0A0L0C1G1 A0A1B0FMX2 A0A194PYL6 A0A1I8NAA4 A0A3B0J1D0 B4LPX7 A0A1W4UZH9 A0A0M4EFQ9 B4J911 B3MG55 A0A2J7QJK2 B4P626 B4HNJ6 B4QBM9 Q6IGN6 B4KR07 B3NSE3 B5E026 B4GAP4 A0A067QWV8 B4MYA6 A0A2P8Y945 A0A0P6H9R9 A0A224XRU3 A0A1D2MKC7 A0A0N7Z9A7 R4G494 A0A0P6BQH1 A0A069DMH8 A6YPT8 E9GMA9 A0A2R7WHZ2 A0A170V1M1 A0A151I8Y4 A0A151WRH4 A0A195B060 A0A158NKU1 A0A226DAL8 A0A151JBG9 A0A195FCK5 T1JGN7 A0A232EIQ7 A0A2L2YAM7 A0A0K8RPH8 E2AC70 A0A023FMX9 B7PSB5 A0A131XEY7 V4B537 A0A2C9JYU2 A0A023G6I7 A0A131YFU9 F0JAE0 L7M5L1 A0A1E1XTR7 A0A0L8GF32 A0A026X231
T1E8A3 A0A2W1BCV6 A0A2M4AYZ8 A0A023ECA3 A0A2M3ZDC0 A0A2M4C5U6 W5JDH7 A0A182FDQ6 Q16HH3 A0A023EDV2 A0A182I3Z5 A0A182V056 A0A182X8W3 A0A182LL31 A0NES5 A0A182YP71 A0A182PFM7 T1E3E8 A0A182NID6 A0A182WC76 A0A182KJ00 A0A1Q3FDD5 A0A182RKG9 B0WMQ2 A0A1B0DMB2 A0A336K369 U5EXY5 A0A1L8DAJ8 A0A1L8EIU9 A0A139WGT4 A0A1B0BKR2 A0A1L8EIZ0 A0A1I8PEU0 A0A1L8EJ21 A0A0L0C1G1 A0A1B0FMX2 A0A194PYL6 A0A1I8NAA4 A0A3B0J1D0 B4LPX7 A0A1W4UZH9 A0A0M4EFQ9 B4J911 B3MG55 A0A2J7QJK2 B4P626 B4HNJ6 B4QBM9 Q6IGN6 B4KR07 B3NSE3 B5E026 B4GAP4 A0A067QWV8 B4MYA6 A0A2P8Y945 A0A0P6H9R9 A0A224XRU3 A0A1D2MKC7 A0A0N7Z9A7 R4G494 A0A0P6BQH1 A0A069DMH8 A6YPT8 E9GMA9 A0A2R7WHZ2 A0A170V1M1 A0A151I8Y4 A0A151WRH4 A0A195B060 A0A158NKU1 A0A226DAL8 A0A151JBG9 A0A195FCK5 T1JGN7 A0A232EIQ7 A0A2L2YAM7 A0A0K8RPH8 E2AC70 A0A023FMX9 B7PSB5 A0A131XEY7 V4B537 A0A2C9JYU2 A0A023G6I7 A0A131YFU9 F0JAE0 L7M5L1 A0A1E1XTR7 A0A0L8GF32 A0A026X231
Ontologies
PANTHER
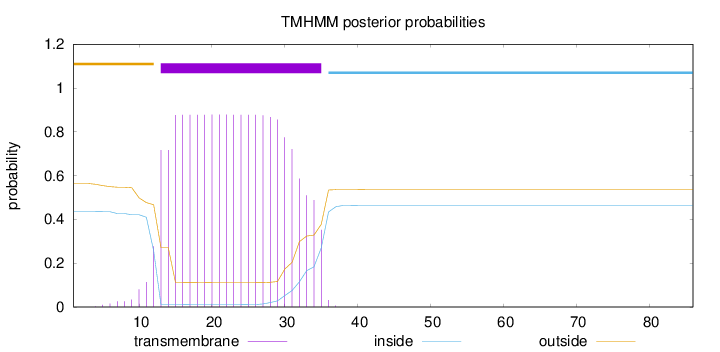
Topology
Subcellular location
Cytoplasm
Length:
86
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.62106
Exp number, first 60 AAs:
18.62106
Total prob of N-in:
0.43603
POSSIBLE N-term signal
sequence
outside
1 - 12
TMhelix
13 - 35
inside
36 - 86
Population Genetic Test Statistics
Pi
253.709326
Theta
172.81806
Tajima's D
1.595579
CLR
0.263295
CSRT
0.808609569521524
Interpretation
Uncertain
