Gene
KWMTBOMO14109 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011203
Annotation
PREDICTED:_glucose-1-phosphatase-like_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 2.221
Sequence
CDS
ATGTTGGCGGTAACTTTTTATTTTTTATTATTATCCGGCGTTGGCTGTCTACACCTCGAGCAAGTATTAATTTTTAGTCGTCACAACATAAGGGTTTCATTTTCAAACCGCATAGATGAATACACGAACAAAATATTTCCGAAATGGTCCAAAGAGCCGGGCCTGTTGACGGAAAAAGGAGCTTTATTGGAAGGCTACATGGGCGAATATCTAACGAAATGGGTGATAGAGAATCAACTCTTACCCGGAACATGTCCCGACAAAGAAACTGTTTTGGTTTATGCGAATAATACGAAACGAACCATAGCTACTGCGAAGGCGTTTGTCGATGCCGCATTCCCTGACTGTAATATAAACGTGAAGTATGAAAAGGATTTTAAAAAAAATGACCTCACCTTTAATTATTATATTCATAACACTACCGAGTCCTACAAACGAAAAGTTGTCGAAGAAATCGAGGAAATGCTCGCCAAATATAAACTAACAGATGCCTACGAAGAATTAGACAAGATTATCGACTTAAAACACTCTAAAAAATGCGAAAGAGAAGGATTCTGCGATTTGGTCCACGATAAGAATATTATACTTTATCACGCTGGCGAGGAACTGGAGCTGAACGGACCTTTGACAATAGGAAATGAAGTAGTTGATAATTTCATAATGAGTTACTACGAAGGACGTCCACTGGAAGAAGTTGCGTGGGGAGAAATCACTAGTCCCGAGCAATGGAAAGTTCTAACTAAAATAACAGCTATGGATGAACAAATACGGTTTAATATTCCCTCAGCTGTCAAGGATACTGCCCGACCTACGATCAGATATTTAAGATCTATATTCACAAATGAAAATATTAAATTTGCATTGATCGTGGGCCACGATGTGAATTTGAACTTAATAACCAACGCTTTAGGTTTCAATCCGTTTGTGATACCTAAGCAGTACGAGCCGTACCCCGTAGGCGGGAAGATTATATTTCAAAAATGGAAGGACGTGGCCGGTGATTATTATTTGAAAGTAGAATACGTTTATCTGTCTTGGGATCAATTAAGAAATGGTGTTAAAATCAACTTTCAAAACCCGCCATTACGAGTTACACTGGGGATCAAAGATTGCGCGATCGATAAAAATGGTTTATGTCCGTGGGATGATTTCATGAAATTAATAAATAAGTATTAA
Protein
MLAVTFYFLLLSGVGCLHLEQVLIFSRHNIRVSFSNRIDEYTNKIFPKWSKEPGLLTEKGALLEGYMGEYLTKWVIENQLLPGTCPDKETVLVYANNTKRTIATAKAFVDAAFPDCNINVKYEKDFKKNDLTFNYYIHNTTESYKRKVVEEIEEMLAKYKLTDAYEELDKIIDLKHSKKCEREGFCDLVHDKNIILYHAGEELELNGPLTIGNEVVDNFIMSYYEGRPLEEVAWGEITSPEQWKVLTKITAMDEQIRFNIPSAVKDTARPTIRYLRSIFTNENIKFALIVGHDVNLNLITNALGFNPFVIPKQYEPYPVGGKIIFQKWKDVAGDYYLKVEYVYLSWDQLRNGVKINFQNPPLRVTLGIKDCAIDKNGLCPWDDFMKLINKY
Summary
Similarity
Belongs to the histidine acid phosphatase family.
Uniprot
H9JNU9
H9JNU7
H9JNU8
H9JNU5
A0A2W1BEE5
A0A2A4IYT5
+ More
A0A194RQR5 A0A0L7K3A8 A0A2W1BNH9 A0A194PYM0 I4DKE4 A0A083ZUA5 A0A2V1H385 A0A3Q0NVX7 A0A2G8A0M1 A0A086GCR1 A0A0P0QGU1 A0A0L7K2Z5 A0A221FRN4 A0A1B3FE38 A0A1Q5W9J2 A0A1C3HG18 A0A1Q4P214 A0A169FMR8 A0A3E2EID8 A0A221DSD7 X2LK33 X2LCN8 A0A3G6CQ42 A0A1C0G629 A0A0J5CUU4 A0A250AW28 A0A2V4G167 A0A3R8TXC9 A0A2V4GQ98 A0A2P8VMC3 A0A085HHH8 A0A3G4SU81 A0A3A5JPQ8 A0A3F3KXR1 A0A3S0APZ7 A0A1B1KTP1 A0A1B7IWJ8 A0A370T804 A0A292AFX6 U2M168 A0A089PUD0 A0A1C7WGT7 A0A0U3SHB9 A0A1B7HR26 A0A1B7HZ46 A0A240C615 A0A1Q5V748 A0A0L7L9U4 A0A3G2IBM1 A0A318P3L6 S3IZW4 A0A3R2D8P6 A0A0X2P5J4 A0A2H1LHW3 A0A379YPH5 S4YL62 A0A2A4IY66 A0A3M3KIM8 A0A0F7HFS7 A0A2W1B8B6 A0A3R2HPC8 A0A3G2FGR8 A0A0X8SLR2 A0A1S8CLE0 A0A3R3CBG4 A0A1W5DM15 A0A2X2GIY9 A0A2X2J3P5 A8GGM4 A0A2X4VEA1 A0A2H1WBS4 A0A085GGY5 A0A0M2K698 A0A381CC18 A0A345CRV5 A0A2J9E7D8 J1GH62 A0A3D5QRT0 A0A2P5IQ07 A0A3S0UCN4 D8MQH9 A0A3A9J2A9 A0A2S7ZZ54 A0A014NIP5 E0LYP8 A0A1B8H7P8 A0A0A3Z568 A0A331L8J7 A0A378FJ87 A0A3M7UIL2 A0A3N4PA42 A0A291E049 D4E497
A0A194RQR5 A0A0L7K3A8 A0A2W1BNH9 A0A194PYM0 I4DKE4 A0A083ZUA5 A0A2V1H385 A0A3Q0NVX7 A0A2G8A0M1 A0A086GCR1 A0A0P0QGU1 A0A0L7K2Z5 A0A221FRN4 A0A1B3FE38 A0A1Q5W9J2 A0A1C3HG18 A0A1Q4P214 A0A169FMR8 A0A3E2EID8 A0A221DSD7 X2LK33 X2LCN8 A0A3G6CQ42 A0A1C0G629 A0A0J5CUU4 A0A250AW28 A0A2V4G167 A0A3R8TXC9 A0A2V4GQ98 A0A2P8VMC3 A0A085HHH8 A0A3G4SU81 A0A3A5JPQ8 A0A3F3KXR1 A0A3S0APZ7 A0A1B1KTP1 A0A1B7IWJ8 A0A370T804 A0A292AFX6 U2M168 A0A089PUD0 A0A1C7WGT7 A0A0U3SHB9 A0A1B7HR26 A0A1B7HZ46 A0A240C615 A0A1Q5V748 A0A0L7L9U4 A0A3G2IBM1 A0A318P3L6 S3IZW4 A0A3R2D8P6 A0A0X2P5J4 A0A2H1LHW3 A0A379YPH5 S4YL62 A0A2A4IY66 A0A3M3KIM8 A0A0F7HFS7 A0A2W1B8B6 A0A3R2HPC8 A0A3G2FGR8 A0A0X8SLR2 A0A1S8CLE0 A0A3R3CBG4 A0A1W5DM15 A0A2X2GIY9 A0A2X2J3P5 A8GGM4 A0A2X4VEA1 A0A2H1WBS4 A0A085GGY5 A0A0M2K698 A0A381CC18 A0A345CRV5 A0A2J9E7D8 J1GH62 A0A3D5QRT0 A0A2P5IQ07 A0A3S0UCN4 D8MQH9 A0A3A9J2A9 A0A2S7ZZ54 A0A014NIP5 E0LYP8 A0A1B8H7P8 A0A0A3Z568 A0A331L8J7 A0A378FJ87 A0A3M7UIL2 A0A3N4PA42 A0A291E049 D4E497
Pubmed
EMBL
BABH01009893
BABH01009897
BABH01009896
BABH01009898
KZ150282
PZC71627.1
+ More
NWSH01004925 PCG64578.1 KQ459875 KPJ19650.1 JTDY01012336 KOB52295.1 KZ149954 PZC76508.1 KQ459593 KPI96250.1 AK401762 BAM18384.1 AYKS02000122 KEY56634.1 PYUJ01000080 PVZ80270.1 LCWI01000001 KKZ20018.1 MSTL01000006 PIJ18962.1 JPUX01000001 KFF88975.1 CP013046 ALL38916.1 JTDY01014338 KOB52039.1 CP018926 ASM12850.1 CP016948 AOF00052.1 LZOB01000011 OKP51878.1 CP021164 PPGF01000001 LT575490 AWQ48708.1 PNU41685.1 SAY43994.1 MJAO01000007 OKB67202.1 FKZE01000001 SAP48203.1 QVOI01000001 RFT81724.1 CP018917 ASL88506.1 KF796611 AHN98126.1 KF796610 AHN98078.1 CP033831 AYZ33663.1 LJDX02000024 OCO27101.1 FCGE01000021 CUZ92713.1 CP014136 ATA18124.1 QJPU01000001 PYA08536.1 QJPV01000001 QMDQ01000009 PYA42071.1 RAU87374.1 PYEP01000002 PSN08713.1 JMPN01000014 KFC95423.1 CP033504 AYU91202.1 QZWH01000027 RJT22235.1 PXZR01000007 PXZP01000003 PXZQ01000001 PXZS01000005 RCEH01000023 RTE96105.1 RTE98319.1 RTF09669.1 RTF13450.1 RTG06826.1 CP015613 ANS43773.1 LXER01000006 OAT34329.1 QRAU01000001 RDL28004.1 CP023956 ATM77910.1 ASZB01000012 ERK10186.1 CP009451 AIR03942.1 LXKR01000001 OCJ46506.1 CP013913 ALX95716.1 LXEP01000032 OAT18098.1 LXEO01000008 OAT20955.1 LT906479 SNW03360.1 MQRH01000033 OKP23014.1 JTDY01002045 KOB72247.1 CP033076 AYN26798.1 PESE01000001 PYD40811.1 ATDT01000009 EPF18041.1 RCUM01000031 MNG49865.1 FBSD01000010 CUW22287.1 LT883155 SMZ57556.1 UGYL01000001 SUI48138.1 CP006566 AGP45185.1 PCG64579.1 RBOX01000004 RMN22483.1 CP011254 AKG72034.1 PZC71628.1 RCUN01000250 MNH75028.1 CP033055 AYM90975.1 CP014017 AMH01410.1 MOXD01000003 OMQ24531.1 RCUE01000002 RCUO01000001 RCUT01002066 RCUN01000004 RCUS01000001 RCUP01012036 MMZ39707.1 MNB54139.1 MND50340.1 MNH51106.1 MNL90746.1 MNV16967.1 FWWG01000043 SMB41287.1 UAUM01000012 SPZ54583.1 UGYN01000002 LR134494 SUI82918.1 VEI71957.1 CP000826 ABV42264.1 LS483469 SQI43610.1 ODYU01007598 SOQ50518.1 JMPI01000022 KFC82980.1 JXNU01000003 KKF34910.1 UIGI01000001 SUW64573.1 CP013970 AXF76172.1 NWFM02000004 PNK63706.1 AKXM01000012 EJF32073.1 PQJV01000005 POW57562.1 RYDU01000001 RTY59907.1 FP236843 CAX59086.1 RARB01000001 RKJ93012.1 PUEK01000002 PQL28524.1 JFHN01000074 EXU73670.1 AEDL01000006 EFM19643.1 LZEY01000045 OBU05095.1 JRUQ01000029 KGT94232.1 UIRA01000025 SVJ53860.1 RHDS01000002 RNA77678.1 RMVG01000017 RPD96373.1 CP023525 ATF93333.1 ADBY01000048 EFE95306.1
NWSH01004925 PCG64578.1 KQ459875 KPJ19650.1 JTDY01012336 KOB52295.1 KZ149954 PZC76508.1 KQ459593 KPI96250.1 AK401762 BAM18384.1 AYKS02000122 KEY56634.1 PYUJ01000080 PVZ80270.1 LCWI01000001 KKZ20018.1 MSTL01000006 PIJ18962.1 JPUX01000001 KFF88975.1 CP013046 ALL38916.1 JTDY01014338 KOB52039.1 CP018926 ASM12850.1 CP016948 AOF00052.1 LZOB01000011 OKP51878.1 CP021164 PPGF01000001 LT575490 AWQ48708.1 PNU41685.1 SAY43994.1 MJAO01000007 OKB67202.1 FKZE01000001 SAP48203.1 QVOI01000001 RFT81724.1 CP018917 ASL88506.1 KF796611 AHN98126.1 KF796610 AHN98078.1 CP033831 AYZ33663.1 LJDX02000024 OCO27101.1 FCGE01000021 CUZ92713.1 CP014136 ATA18124.1 QJPU01000001 PYA08536.1 QJPV01000001 QMDQ01000009 PYA42071.1 RAU87374.1 PYEP01000002 PSN08713.1 JMPN01000014 KFC95423.1 CP033504 AYU91202.1 QZWH01000027 RJT22235.1 PXZR01000007 PXZP01000003 PXZQ01000001 PXZS01000005 RCEH01000023 RTE96105.1 RTE98319.1 RTF09669.1 RTF13450.1 RTG06826.1 CP015613 ANS43773.1 LXER01000006 OAT34329.1 QRAU01000001 RDL28004.1 CP023956 ATM77910.1 ASZB01000012 ERK10186.1 CP009451 AIR03942.1 LXKR01000001 OCJ46506.1 CP013913 ALX95716.1 LXEP01000032 OAT18098.1 LXEO01000008 OAT20955.1 LT906479 SNW03360.1 MQRH01000033 OKP23014.1 JTDY01002045 KOB72247.1 CP033076 AYN26798.1 PESE01000001 PYD40811.1 ATDT01000009 EPF18041.1 RCUM01000031 MNG49865.1 FBSD01000010 CUW22287.1 LT883155 SMZ57556.1 UGYL01000001 SUI48138.1 CP006566 AGP45185.1 PCG64579.1 RBOX01000004 RMN22483.1 CP011254 AKG72034.1 PZC71628.1 RCUN01000250 MNH75028.1 CP033055 AYM90975.1 CP014017 AMH01410.1 MOXD01000003 OMQ24531.1 RCUE01000002 RCUO01000001 RCUT01002066 RCUN01000004 RCUS01000001 RCUP01012036 MMZ39707.1 MNB54139.1 MND50340.1 MNH51106.1 MNL90746.1 MNV16967.1 FWWG01000043 SMB41287.1 UAUM01000012 SPZ54583.1 UGYN01000002 LR134494 SUI82918.1 VEI71957.1 CP000826 ABV42264.1 LS483469 SQI43610.1 ODYU01007598 SOQ50518.1 JMPI01000022 KFC82980.1 JXNU01000003 KKF34910.1 UIGI01000001 SUW64573.1 CP013970 AXF76172.1 NWFM02000004 PNK63706.1 AKXM01000012 EJF32073.1 PQJV01000005 POW57562.1 RYDU01000001 RTY59907.1 FP236843 CAX59086.1 RARB01000001 RKJ93012.1 PUEK01000002 PQL28524.1 JFHN01000074 EXU73670.1 AEDL01000006 EFM19643.1 LZEY01000045 OBU05095.1 JRUQ01000029 KGT94232.1 UIRA01000025 SVJ53860.1 RHDS01000002 RNA77678.1 RMVG01000017 RPD96373.1 CP023525 ATF93333.1 ADBY01000048 EFE95306.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000053268
UP000017810
+ More
UP000244888 UP000034361 UP000229306 UP000028708 UP000058492 UP000198289 UP000077316 UP000185968 UP000236556 UP000246645 UP000185770 UP000078180 UP000261108 UP000198428 UP000282317 UP000050507 UP000054899 UP000217182 UP000248086 UP000247670 UP000251655 UP000240212 UP000028624 UP000281251 UP000276295 UP000275654 UP000287517 UP000287538 UP000288290 UP000288299 UP000013567 UP000078410 UP000255042 UP000221800 UP000016623 UP000029481 UP000093001 UP000061771 UP000078504 UP000078286 UP000215134 UP000186436 UP000268079 UP000248196 UP000014585 UP000242817 UP000246092 UP000254742 UP000014900 UP000034699 UP000281837 UP000062490 UP000216021 UP000192912 UP000250791 UP000255529 UP000273164 UP000007074 UP000248897 UP000028653 UP000033924 UP000255528 UP000264980 UP000218868 UP000005486 UP000246545 UP000286884 UP000008793 UP000275729 UP000238919 UP000019918 UP000004279 UP000092377 UP000030351 UP000258067 UP000278924 UP000281332 UP000217979 UP000005723
UP000244888 UP000034361 UP000229306 UP000028708 UP000058492 UP000198289 UP000077316 UP000185968 UP000236556 UP000246645 UP000185770 UP000078180 UP000261108 UP000198428 UP000282317 UP000050507 UP000054899 UP000217182 UP000248086 UP000247670 UP000251655 UP000240212 UP000028624 UP000281251 UP000276295 UP000275654 UP000287517 UP000287538 UP000288290 UP000288299 UP000013567 UP000078410 UP000255042 UP000221800 UP000016623 UP000029481 UP000093001 UP000061771 UP000078504 UP000078286 UP000215134 UP000186436 UP000268079 UP000248196 UP000014585 UP000242817 UP000246092 UP000254742 UP000014900 UP000034699 UP000281837 UP000062490 UP000216021 UP000192912 UP000250791 UP000255529 UP000273164 UP000007074 UP000248897 UP000028653 UP000033924 UP000255528 UP000264980 UP000218868 UP000005486 UP000246545 UP000286884 UP000008793 UP000275729 UP000238919 UP000019918 UP000004279 UP000092377 UP000030351 UP000258067 UP000278924 UP000281332 UP000217979 UP000005723
Pfam
PF00328 His_Phos_2
SUPFAM
SSF53254
SSF53254
Gene 3D
ProteinModelPortal
H9JNU9
H9JNU7
H9JNU8
H9JNU5
A0A2W1BEE5
A0A2A4IYT5
+ More
A0A194RQR5 A0A0L7K3A8 A0A2W1BNH9 A0A194PYM0 I4DKE4 A0A083ZUA5 A0A2V1H385 A0A3Q0NVX7 A0A2G8A0M1 A0A086GCR1 A0A0P0QGU1 A0A0L7K2Z5 A0A221FRN4 A0A1B3FE38 A0A1Q5W9J2 A0A1C3HG18 A0A1Q4P214 A0A169FMR8 A0A3E2EID8 A0A221DSD7 X2LK33 X2LCN8 A0A3G6CQ42 A0A1C0G629 A0A0J5CUU4 A0A250AW28 A0A2V4G167 A0A3R8TXC9 A0A2V4GQ98 A0A2P8VMC3 A0A085HHH8 A0A3G4SU81 A0A3A5JPQ8 A0A3F3KXR1 A0A3S0APZ7 A0A1B1KTP1 A0A1B7IWJ8 A0A370T804 A0A292AFX6 U2M168 A0A089PUD0 A0A1C7WGT7 A0A0U3SHB9 A0A1B7HR26 A0A1B7HZ46 A0A240C615 A0A1Q5V748 A0A0L7L9U4 A0A3G2IBM1 A0A318P3L6 S3IZW4 A0A3R2D8P6 A0A0X2P5J4 A0A2H1LHW3 A0A379YPH5 S4YL62 A0A2A4IY66 A0A3M3KIM8 A0A0F7HFS7 A0A2W1B8B6 A0A3R2HPC8 A0A3G2FGR8 A0A0X8SLR2 A0A1S8CLE0 A0A3R3CBG4 A0A1W5DM15 A0A2X2GIY9 A0A2X2J3P5 A8GGM4 A0A2X4VEA1 A0A2H1WBS4 A0A085GGY5 A0A0M2K698 A0A381CC18 A0A345CRV5 A0A2J9E7D8 J1GH62 A0A3D5QRT0 A0A2P5IQ07 A0A3S0UCN4 D8MQH9 A0A3A9J2A9 A0A2S7ZZ54 A0A014NIP5 E0LYP8 A0A1B8H7P8 A0A0A3Z568 A0A331L8J7 A0A378FJ87 A0A3M7UIL2 A0A3N4PA42 A0A291E049 D4E497
A0A194RQR5 A0A0L7K3A8 A0A2W1BNH9 A0A194PYM0 I4DKE4 A0A083ZUA5 A0A2V1H385 A0A3Q0NVX7 A0A2G8A0M1 A0A086GCR1 A0A0P0QGU1 A0A0L7K2Z5 A0A221FRN4 A0A1B3FE38 A0A1Q5W9J2 A0A1C3HG18 A0A1Q4P214 A0A169FMR8 A0A3E2EID8 A0A221DSD7 X2LK33 X2LCN8 A0A3G6CQ42 A0A1C0G629 A0A0J5CUU4 A0A250AW28 A0A2V4G167 A0A3R8TXC9 A0A2V4GQ98 A0A2P8VMC3 A0A085HHH8 A0A3G4SU81 A0A3A5JPQ8 A0A3F3KXR1 A0A3S0APZ7 A0A1B1KTP1 A0A1B7IWJ8 A0A370T804 A0A292AFX6 U2M168 A0A089PUD0 A0A1C7WGT7 A0A0U3SHB9 A0A1B7HR26 A0A1B7HZ46 A0A240C615 A0A1Q5V748 A0A0L7L9U4 A0A3G2IBM1 A0A318P3L6 S3IZW4 A0A3R2D8P6 A0A0X2P5J4 A0A2H1LHW3 A0A379YPH5 S4YL62 A0A2A4IY66 A0A3M3KIM8 A0A0F7HFS7 A0A2W1B8B6 A0A3R2HPC8 A0A3G2FGR8 A0A0X8SLR2 A0A1S8CLE0 A0A3R3CBG4 A0A1W5DM15 A0A2X2GIY9 A0A2X2J3P5 A8GGM4 A0A2X4VEA1 A0A2H1WBS4 A0A085GGY5 A0A0M2K698 A0A381CC18 A0A345CRV5 A0A2J9E7D8 J1GH62 A0A3D5QRT0 A0A2P5IQ07 A0A3S0UCN4 D8MQH9 A0A3A9J2A9 A0A2S7ZZ54 A0A014NIP5 E0LYP8 A0A1B8H7P8 A0A0A3Z568 A0A331L8J7 A0A378FJ87 A0A3M7UIL2 A0A3N4PA42 A0A291E049 D4E497
PDB
1NT4
E-value=2.18615e-80,
Score=761
Ontologies
GO
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.876766
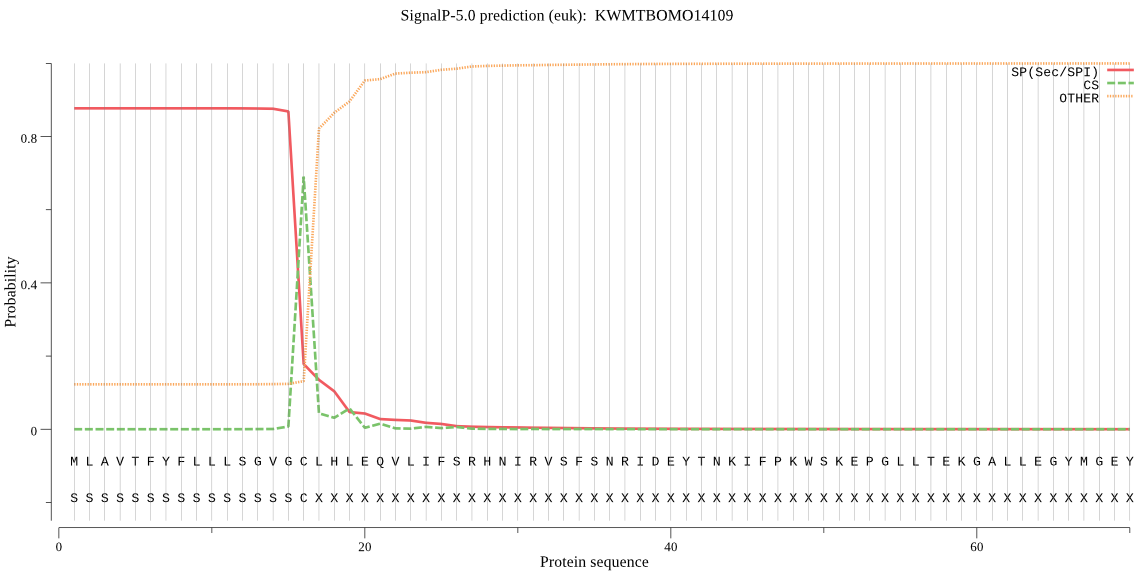
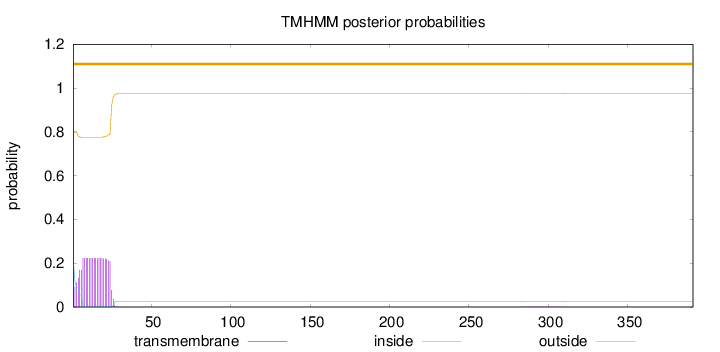
Length:
391
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.82379
Exp number, first 60 AAs:
4.79169
Total prob of N-in:
0.20140
outside
1 - 391
Population Genetic Test Statistics
Pi
395.306447
Theta
196.413702
Tajima's D
3.14965
CLR
0
CSRT
0.985150742462877
Interpretation
Uncertain
