Gene
KWMTBOMO14105
Pre Gene Modal
BGIBMGA011390
Annotation
PREDICTED:_P_protein-like_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.899
Sequence
CDS
ATGCGCAAGACGAGCACTTCCGCCTACGTAAAGCACTTGATAAAAATCTCTATACTCCTGGCATGCTGGATGTTCTTCACGATCGTCTTCATGATGTACAATGAGAAGGTGGAAGTGTTCATGAGCTCGTCCGTGGCTCCGGGAGAGGTCAAGAGCTACATCATAGACTTAAAAGAGGACAAGCTATCATTGTCCTTAAGCCTTTCTGGGCCGTTCCTCTCGGAGCAGTGGCAGGCCAAGTTGAATTCCACGGAGGGAGTAAAGAAGATGGAGGTTTGGCTTGAAAGGCGGCCCGATGATGACAGCAGAGGAGTATCGGTCGAGATATCAAAGACATGGGAAATAGTATTGCTAGAGAACGAAACGGATTTCTGTGAGGGCGAACTGAGGTCGAACGTTCTAGAATTGAAGACGTTGAACGCAACCAATAATGGGTCTTACGCTGTCGCGATAAGAACTAACGCGAACGAGACGATACCGTTCCAAATGAGCTACAAGATTGAGCCGCTGGATCAGACTTCGGGAGTGATTTACGCTTCAATTTTACTGCTGGGGTTGTATGTTCTGATAATATTTGAGATCATAAACCGAACCATGGCAGCAGTTATCCTGTCAACAACGGCCATGGCCGTCCTCTCGTTACTAGGCGAAAGGCCTTCCTTGCCAGAATTGATCTCGTGGCTGGATATGGAGACGTTGCTGCTGCTCTTTAGCATGATGCTGCTTGTAGCGATCATGGCTGAGACGGGACTGTTCGACTATCTCGCCGTCTACACATTTGAGGTTACAAGAGGGAAGATCTGGCCCCTGATCTCCGTTCTGTGCGGAATAACGGCGATGCTATCAACATTTTTAGACAACGTTACAACTGTATTACTAATGACACCGGTCACAATAAGGTTATGTGAAGTGATGGATATGGATCCTATTCCCGTACTGATGTCCATGGTCATATACAGTAATATTGGTGGGACCGCTACTCCTGTGGGCGATCCGCCCAACGTTATAATAGCTTCAAATAAAGCTGTTAGTCAAGCTGGGATAAACTTCACGAATTTCACATTGCACATGGCACTGGGAATCGTCTTCGTGTGTCTGCAGACCTACGTTCAATTGAGATTCATTTATCGAGACACGAATAGGCTACGTTTAAACGAACCAAGAGAGATACAAGATCTCCGCCAACAAATCTCGATATGGCGTCGCGCCGCCGACTCCCTCCCGCATCTCAGTAGAGACGAACAGGTGGTCAGAGAGAGACTCGACAAGAAGATACGTAAATTGACTTCCCAGTTGGATCTTTTCGTTAGGGAGAGTAAAAAGAGAGCCTGCCCTAAGACCACATTCAGGAGCACCTTGGCCGAGATGAAAGAGAAGTACAAGATTCGCAATAAGCCGTTATTGGCGAAATCTTCAGTGGCCATCGGTTTCGTCGTGCTTGTGTTCTTCTTGCACTCCATACCGGAATTCAATCGCGTGTCCCTGGGCTGGACGGCGTTGCTGGGCGCGATCCTGCTGCTGATCCTGGCGGACCGCGAGGACCTGGAGCCGGTGCTGCACCGAGTCGAGTGGTCCACACTGCTGTTCTTCGCGGCACTCTTTGTTTTTATGGAGGCCCTCTCGAAGTTGGGGCTCATCGTTTTCATTGGAGGCCTCACGGAGTCCATCATTCTCAAGGTGGACGAAAGTGCTAGACTGGCCGTGGCGATACTGCTTATTTTGTGGGTGTCAGGTTTGACGTCGGCCTTCGTAGACAACATCCCGCTGACGACCATGATGATCAGAGTGGTGACCTCGCTCGGGTCGAACCCGAACCTCAATCTTCCGATGCAGCCGCTGATCTGGGCACTTTGCTACGGAGCTTGTCTAGGAGGTAACGGTACCCTGATAGGCGCGAGCGCCAACGTGGTCTGCGCTGGCGTGGCTGAGCAACACGGATACAAGTTCACCTTCATGAACTTCTTCAAAATAGGCTTCCCGATCATGATAGGCCACTTGGTCGTAGCCTCACTGTACCTGATTATATGCCACTGCGTTTTTGCCTGGCATTGA
Protein
MRKTSTSAYVKHLIKISILLACWMFFTIVFMMYNEKVEVFMSSSVAPGEVKSYIIDLKEDKLSLSLSLSGPFLSEQWQAKLNSTEGVKKMEVWLERRPDDDSRGVSVEISKTWEIVLLENETDFCEGELRSNVLELKTLNATNNGSYAVAIRTNANETIPFQMSYKIEPLDQTSGVIYASILLLGLYVLIIFEIINRTMAAVILSTTAMAVLSLLGERPSLPELISWLDMETLLLLFSMMLLVAIMAETGLFDYLAVYTFEVTRGKIWPLISVLCGITAMLSTFLDNVTTVLLMTPVTIRLCEVMDMDPIPVLMSMVIYSNIGGTATPVGDPPNVIIASNKAVSQAGINFTNFTLHMALGIVFVCLQTYVQLRFIYRDTNRLRLNEPREIQDLRQQISIWRRAADSLPHLSRDEQVVRERLDKKIRKLTSQLDLFVRESKKRACPKTTFRSTLAEMKEKYKIRNKPLLAKSSVAIGFVVLVFFLHSIPEFNRVSLGWTALLGAILLLILADREDLEPVLHRVEWSTLLFFAALFVFMEALSKLGLIVFIGGLTESIILKVDESARLAVAILLILWVSGLTSAFVDNIPLTTMMIRVVTSLGSNPNLNLPMQPLIWALCYGACLGGNGTLIGASANVVCAGVAEQHGYKFTFMNFFKIGFPIMIGHLVVASLYLIICHCVFAWH
Summary
Uniprot
H9JPD6
A0A2H1W219
A0A3S2NV69
A0A2A4JQX1
A0A194PYP6
A0A194RQT6
+ More
A0A2W1B5T8 A0A2A4JVQ7 A0A2W1BCA3 U5EQX7 A0A1L8DEW3 A0A194PU37 A0A194PTH2 A0A182PG17 A0A182Q2A0 A0A182F4R7 A0A182VUI4 A0A182NU64 W5J9Z5 Q7PVG5 A0A182X0R7 A0A1S4H0M1 A0A182HGF4 A0A182L6G4 A0A182GEX5 A0A182V329 A0A182YDN9 A0A182RAX9 A0A182MMN2 A0A182K975 A0A023EW65 A0A182TF71 A0A336LQF6 A0A232EZM2 A0A1J1IKS2 A0A1Q3FEZ3 A0A1Q3FEZ7 A0A1Q3FEV8 A0A1Q3FEP7 A0A1Q3FES9 A0A0N8ERX9 A0A182J9C5 A0A154NVS8 K7J3V8 Q170B8 A0A034V4S7 A0A0K8W422 A0A0A1WYG4 A0A0A1WJ94 W8AKI0 T1EAR5 B4JCQ7 T1PFL4 A0A088A3Y6 A0A067RB00 A0A1W4UI99 A0A1W4UW27 A0A1I8MTA9 A0A1W4UXH3 A0A1I8MTB5 A0A0Q9WX89 E2AJY6 B4MDJ4 A0A0Q9WM04 A0A0J9QWS6 B4Q345 A0A0J9TFR0 B4G9J0 B4I182 Q8IGX6 Q9Y163 Q8I930 A0A0R3NSS9 A0A0R1DPW4 A0A0Q5WGC6 A0A0Q5W9B1 Q9VR47 A0A1I8Q160 A0A0R1DRT0 A0A0M3QU56 A0A0R3NSJ7 A0A1I8Q130 B4NWB0 B3N4B6 A0A3B0K2C5 A0A2J7QQX2 A0A3B0JUJ9 Q29MA2 A0A1I8Q162 A0A0Q9X4F0 B4KJ44 A0A0P8XHQ7 A0A0Q9X3R9 A0A0P9A7N2 A0A0J9QVU0 A0A1B0FJS8 B3MVJ9 E0VTX1 A0A0Q9X1N6
A0A2W1B5T8 A0A2A4JVQ7 A0A2W1BCA3 U5EQX7 A0A1L8DEW3 A0A194PU37 A0A194PTH2 A0A182PG17 A0A182Q2A0 A0A182F4R7 A0A182VUI4 A0A182NU64 W5J9Z5 Q7PVG5 A0A182X0R7 A0A1S4H0M1 A0A182HGF4 A0A182L6G4 A0A182GEX5 A0A182V329 A0A182YDN9 A0A182RAX9 A0A182MMN2 A0A182K975 A0A023EW65 A0A182TF71 A0A336LQF6 A0A232EZM2 A0A1J1IKS2 A0A1Q3FEZ3 A0A1Q3FEZ7 A0A1Q3FEV8 A0A1Q3FEP7 A0A1Q3FES9 A0A0N8ERX9 A0A182J9C5 A0A154NVS8 K7J3V8 Q170B8 A0A034V4S7 A0A0K8W422 A0A0A1WYG4 A0A0A1WJ94 W8AKI0 T1EAR5 B4JCQ7 T1PFL4 A0A088A3Y6 A0A067RB00 A0A1W4UI99 A0A1W4UW27 A0A1I8MTA9 A0A1W4UXH3 A0A1I8MTB5 A0A0Q9WX89 E2AJY6 B4MDJ4 A0A0Q9WM04 A0A0J9QWS6 B4Q345 A0A0J9TFR0 B4G9J0 B4I182 Q8IGX6 Q9Y163 Q8I930 A0A0R3NSS9 A0A0R1DPW4 A0A0Q5WGC6 A0A0Q5W9B1 Q9VR47 A0A1I8Q160 A0A0R1DRT0 A0A0M3QU56 A0A0R3NSJ7 A0A1I8Q130 B4NWB0 B3N4B6 A0A3B0K2C5 A0A2J7QQX2 A0A3B0JUJ9 Q29MA2 A0A1I8Q162 A0A0Q9X4F0 B4KJ44 A0A0P8XHQ7 A0A0Q9X3R9 A0A0P9A7N2 A0A0J9QVU0 A0A1B0FJS8 B3MVJ9 E0VTX1 A0A0Q9X1N6
Pubmed
19121390
26354079
28756777
20920257
23761445
12364791
+ More
20966253 26483478 25244985 24945155 28648823 26999592 20075255 17510324 25348373 25830018 24495485 17994087 24845553 25315136 18057021 20798317 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 23185243 20566863
20966253 26483478 25244985 24945155 28648823 26999592 20075255 17510324 25348373 25830018 24495485 17994087 24845553 25315136 18057021 20798317 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 23185243 20566863
EMBL
BABH01009882
BABH01009883
ODYU01005849
SOQ47140.1
RSAL01000061
RVE49595.1
+ More
NWSH01000786 PCG74209.1 KQ459593 KPI96275.1 KQ459875 KPJ19670.1 KZ150482 PZC70721.1 NWSH01000477 PCG76127.1 KZ150171 PZC72608.1 GANO01004039 JAB55832.1 GFDF01009081 JAV05003.1 KPI96269.1 KPI96273.1 AXCN02000773 ADMH02001962 ETN60233.1 AAAB01008984 EAA14953.3 APCN01004223 JXUM01060995 JXUM01060996 KQ562130 KXJ76619.1 AXCM01000685 GAPW01000206 JAC13392.1 UFQT01000114 SSX20252.1 NNAY01001501 OXU23779.1 CVRI01000054 CRL00831.1 GFDL01008953 JAV26092.1 GFDL01008936 JAV26109.1 GFDL01008962 JAV26083.1 GFDL01009032 JAV26013.1 GFDL01008994 JAV26051.1 GDUN01001018 JAN94901.1 KQ434769 KZC03779.1 CH477477 EAT40274.1 GAKP01022216 GAKP01022212 JAC36740.1 GDHF01018577 GDHF01006502 GDHF01005578 GDHF01004669 JAI33737.1 JAI45812.1 JAI46736.1 JAI47645.1 GBXI01010849 JAD03443.1 GBXI01015385 JAC98906.1 GAMC01021322 JAB85233.1 GAMD01000808 JAB00783.1 CH916368 EDW04221.1 KA646955 AFP61584.1 KK852579 KDR20906.1 CH940661 KRF85587.1 KRF85588.1 GL440100 EFN66302.1 EDW71255.1 KRF85586.1 CM002910 KMY88169.1 CM000361 EDX03761.1 KMY88162.1 KMY88165.1 CH479180 EDW29020.1 CH480820 EDW54289.1 BT001540 AAN71295.1 AF145605 AE014134 AY050572 AAD38580.1 AAF50961.1 AAL11434.1 AY050571 BT016131 AAF50960.3 AAL11433.1 AAO41154.1 AAO41155.1 AAO41156.1 AAV37016.1 AGB92596.1 CH379060 KRT04194.1 CM000157 KRJ97778.1 CH954177 KQS70133.1 KQS70134.1 BT044574 AAF50959.2 ACI16536.1 AHN54122.1 KRJ97776.1 KRJ97777.1 CP012523 ALC40049.1 KRT04193.1 KRT04195.1 EDW88427.1 EDV57786.1 OUUW01000004 SPP79151.1 NEVH01012082 PNF30980.1 SPP79150.1 EAL33792.1 CH933807 KRG02677.1 EDW11406.1 CH902624 KPU74378.1 KRG02678.1 KPU74379.1 KMY88167.1 CCAG010022036 EDV33264.2 DS235774 EEB16827.1 CH963920 KRF98706.1
NWSH01000786 PCG74209.1 KQ459593 KPI96275.1 KQ459875 KPJ19670.1 KZ150482 PZC70721.1 NWSH01000477 PCG76127.1 KZ150171 PZC72608.1 GANO01004039 JAB55832.1 GFDF01009081 JAV05003.1 KPI96269.1 KPI96273.1 AXCN02000773 ADMH02001962 ETN60233.1 AAAB01008984 EAA14953.3 APCN01004223 JXUM01060995 JXUM01060996 KQ562130 KXJ76619.1 AXCM01000685 GAPW01000206 JAC13392.1 UFQT01000114 SSX20252.1 NNAY01001501 OXU23779.1 CVRI01000054 CRL00831.1 GFDL01008953 JAV26092.1 GFDL01008936 JAV26109.1 GFDL01008962 JAV26083.1 GFDL01009032 JAV26013.1 GFDL01008994 JAV26051.1 GDUN01001018 JAN94901.1 KQ434769 KZC03779.1 CH477477 EAT40274.1 GAKP01022216 GAKP01022212 JAC36740.1 GDHF01018577 GDHF01006502 GDHF01005578 GDHF01004669 JAI33737.1 JAI45812.1 JAI46736.1 JAI47645.1 GBXI01010849 JAD03443.1 GBXI01015385 JAC98906.1 GAMC01021322 JAB85233.1 GAMD01000808 JAB00783.1 CH916368 EDW04221.1 KA646955 AFP61584.1 KK852579 KDR20906.1 CH940661 KRF85587.1 KRF85588.1 GL440100 EFN66302.1 EDW71255.1 KRF85586.1 CM002910 KMY88169.1 CM000361 EDX03761.1 KMY88162.1 KMY88165.1 CH479180 EDW29020.1 CH480820 EDW54289.1 BT001540 AAN71295.1 AF145605 AE014134 AY050572 AAD38580.1 AAF50961.1 AAL11434.1 AY050571 BT016131 AAF50960.3 AAL11433.1 AAO41154.1 AAO41155.1 AAO41156.1 AAV37016.1 AGB92596.1 CH379060 KRT04194.1 CM000157 KRJ97778.1 CH954177 KQS70133.1 KQS70134.1 BT044574 AAF50959.2 ACI16536.1 AHN54122.1 KRJ97776.1 KRJ97777.1 CP012523 ALC40049.1 KRT04193.1 KRT04195.1 EDW88427.1 EDV57786.1 OUUW01000004 SPP79151.1 NEVH01012082 PNF30980.1 SPP79150.1 EAL33792.1 CH933807 KRG02677.1 EDW11406.1 CH902624 KPU74378.1 KRG02678.1 KPU74379.1 KMY88167.1 CCAG010022036 EDV33264.2 DS235774 EEB16827.1 CH963920 KRF98706.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000075885
+ More
UP000075886 UP000069272 UP000075920 UP000075884 UP000000673 UP000007062 UP000076407 UP000075840 UP000075882 UP000069940 UP000249989 UP000075903 UP000076408 UP000075900 UP000075883 UP000075881 UP000075902 UP000215335 UP000183832 UP000075880 UP000076502 UP000002358 UP000008820 UP000001070 UP000005203 UP000027135 UP000192221 UP000095301 UP000008792 UP000000311 UP000000304 UP000008744 UP000001292 UP000000803 UP000001819 UP000002282 UP000008711 UP000095300 UP000092553 UP000268350 UP000235965 UP000009192 UP000007801 UP000092444 UP000009046 UP000007798
UP000075886 UP000069272 UP000075920 UP000075884 UP000000673 UP000007062 UP000076407 UP000075840 UP000075882 UP000069940 UP000249989 UP000075903 UP000076408 UP000075900 UP000075883 UP000075881 UP000075902 UP000215335 UP000183832 UP000075880 UP000076502 UP000002358 UP000008820 UP000001070 UP000005203 UP000027135 UP000192221 UP000095301 UP000008792 UP000000311 UP000000304 UP000008744 UP000001292 UP000000803 UP000001819 UP000002282 UP000008711 UP000095300 UP000092553 UP000268350 UP000235965 UP000009192 UP000007801 UP000092444 UP000009046 UP000007798
Pfam
PF03600 CitMHS
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JPD6
A0A2H1W219
A0A3S2NV69
A0A2A4JQX1
A0A194PYP6
A0A194RQT6
+ More
A0A2W1B5T8 A0A2A4JVQ7 A0A2W1BCA3 U5EQX7 A0A1L8DEW3 A0A194PU37 A0A194PTH2 A0A182PG17 A0A182Q2A0 A0A182F4R7 A0A182VUI4 A0A182NU64 W5J9Z5 Q7PVG5 A0A182X0R7 A0A1S4H0M1 A0A182HGF4 A0A182L6G4 A0A182GEX5 A0A182V329 A0A182YDN9 A0A182RAX9 A0A182MMN2 A0A182K975 A0A023EW65 A0A182TF71 A0A336LQF6 A0A232EZM2 A0A1J1IKS2 A0A1Q3FEZ3 A0A1Q3FEZ7 A0A1Q3FEV8 A0A1Q3FEP7 A0A1Q3FES9 A0A0N8ERX9 A0A182J9C5 A0A154NVS8 K7J3V8 Q170B8 A0A034V4S7 A0A0K8W422 A0A0A1WYG4 A0A0A1WJ94 W8AKI0 T1EAR5 B4JCQ7 T1PFL4 A0A088A3Y6 A0A067RB00 A0A1W4UI99 A0A1W4UW27 A0A1I8MTA9 A0A1W4UXH3 A0A1I8MTB5 A0A0Q9WX89 E2AJY6 B4MDJ4 A0A0Q9WM04 A0A0J9QWS6 B4Q345 A0A0J9TFR0 B4G9J0 B4I182 Q8IGX6 Q9Y163 Q8I930 A0A0R3NSS9 A0A0R1DPW4 A0A0Q5WGC6 A0A0Q5W9B1 Q9VR47 A0A1I8Q160 A0A0R1DRT0 A0A0M3QU56 A0A0R3NSJ7 A0A1I8Q130 B4NWB0 B3N4B6 A0A3B0K2C5 A0A2J7QQX2 A0A3B0JUJ9 Q29MA2 A0A1I8Q162 A0A0Q9X4F0 B4KJ44 A0A0P8XHQ7 A0A0Q9X3R9 A0A0P9A7N2 A0A0J9QVU0 A0A1B0FJS8 B3MVJ9 E0VTX1 A0A0Q9X1N6
A0A2W1B5T8 A0A2A4JVQ7 A0A2W1BCA3 U5EQX7 A0A1L8DEW3 A0A194PU37 A0A194PTH2 A0A182PG17 A0A182Q2A0 A0A182F4R7 A0A182VUI4 A0A182NU64 W5J9Z5 Q7PVG5 A0A182X0R7 A0A1S4H0M1 A0A182HGF4 A0A182L6G4 A0A182GEX5 A0A182V329 A0A182YDN9 A0A182RAX9 A0A182MMN2 A0A182K975 A0A023EW65 A0A182TF71 A0A336LQF6 A0A232EZM2 A0A1J1IKS2 A0A1Q3FEZ3 A0A1Q3FEZ7 A0A1Q3FEV8 A0A1Q3FEP7 A0A1Q3FES9 A0A0N8ERX9 A0A182J9C5 A0A154NVS8 K7J3V8 Q170B8 A0A034V4S7 A0A0K8W422 A0A0A1WYG4 A0A0A1WJ94 W8AKI0 T1EAR5 B4JCQ7 T1PFL4 A0A088A3Y6 A0A067RB00 A0A1W4UI99 A0A1W4UW27 A0A1I8MTA9 A0A1W4UXH3 A0A1I8MTB5 A0A0Q9WX89 E2AJY6 B4MDJ4 A0A0Q9WM04 A0A0J9QWS6 B4Q345 A0A0J9TFR0 B4G9J0 B4I182 Q8IGX6 Q9Y163 Q8I930 A0A0R3NSS9 A0A0R1DPW4 A0A0Q5WGC6 A0A0Q5W9B1 Q9VR47 A0A1I8Q160 A0A0R1DRT0 A0A0M3QU56 A0A0R3NSJ7 A0A1I8Q130 B4NWB0 B3N4B6 A0A3B0K2C5 A0A2J7QQX2 A0A3B0JUJ9 Q29MA2 A0A1I8Q162 A0A0Q9X4F0 B4KJ44 A0A0P8XHQ7 A0A0Q9X3R9 A0A0P9A7N2 A0A0J9QVU0 A0A1B0FJS8 B3MVJ9 E0VTX1 A0A0Q9X1N6
PDB
4F35
E-value=2.14213e-07,
Score=134
Ontologies
GO
Topology
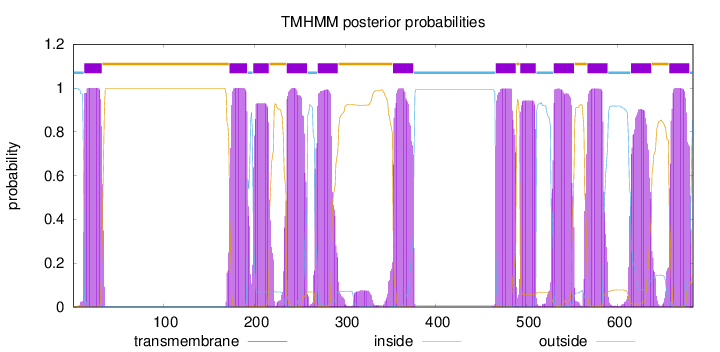
Length:
683
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
254.57012
Exp number, first 60 AAs:
21.12431
Total prob of N-in:
0.99800
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 32
outside
33 - 172
TMhelix
173 - 192
inside
193 - 198
TMhelix
199 - 216
outside
217 - 235
TMhelix
236 - 258
inside
259 - 269
TMhelix
270 - 292
outside
293 - 352
TMhelix
353 - 375
inside
376 - 465
TMhelix
466 - 488
outside
489 - 492
TMhelix
493 - 510
inside
511 - 529
TMhelix
530 - 552
outside
553 - 566
TMhelix
567 - 589
inside
590 - 614
TMhelix
615 - 637
outside
638 - 656
TMhelix
657 - 679
inside
680 - 683
Population Genetic Test Statistics
Pi
204.735691
Theta
159.326167
Tajima's D
1.119002
CLR
0.454677
CSRT
0.695765211739413
Interpretation
Uncertain
