Gene
KWMTBOMO14099 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011387
Annotation
PREDICTED:_lachesin_[Amyelois_transitella]
Full name
Lachesin
Location in the cell
PlasmaMembrane Reliability : 1.562
Sequence
CDS
ATGGATCGAAATATTTACGTTTACGTTTTTTACACTTTACTATTGTATTTCACTTGTAATGTATCAGCCCAACGGACACCAACAATCTCGCACATCACACAAGAGCAGATCAGGGATATCGGAGGCCAGGTAGACCTGGAATGTTCCGTGCATTATGCACAAGAGTTTCCTGTTGTCTGGACCAAACGCGATCGTTTGAAGACGATCGAATCGATACCCCTGTCGACCAACGCCGGGCTCATCATCAAGGACTCTAGGTTCTCGCTGCGCCACGACGAGGCAACGGCCACGTTTACGCTCTCCATCAGGGATATTCAAGAGAACGACGCCGGCTGGTACCAGTGCCAAGTGCTGATGTCCATAAGCAATAAGATCGCCGCTGAAGTGGAGCTCCAGGTCAGGAGGCCCCCCATTATATCGGACAATTCCACCAGGTCAATCGTCGCTAGTGAAGGGGAAAGTGTTCAAATGGAATGCTACGCTGGTGGCTTCCCGCCCCCGAAGATATCCTGGCGCCGCGAGAACAACGCCATCCTGCCGACGGGCGGCTCCATCTACAGGGGCAACGTGCTGAAGATAAGCGCCGTCCACAAGGAGGATCGTGGCACTTATTACTGCGTGGCGGACAATGGCGTCGGCAAAGGCGCCCGTCGCAACATCAACCTCGAGGTGGAATTCGCGCCCGTCATCTCCGTGCCCCGCCCGCGCCTCTCCCAGGCCCTCCAATACGACATGGACCTGGAGTGCCACGTGGAGGCCTACCCGCCGCCTGCCATCACCTGGCTGAAAGATGGCATCCAGCTTTCGAACAACCAACATTATAGGATCTCGCATTTCGCAACGGCGGACGAGTTCACGGACACCACGATCAGGGTTATCACGATCGAGAAGCGACAGTACGGGCTGTTCACGTGCAAAGCTCAGAATAAGCTGGGCACCGCGGAAGGGAACGTCGAGCTCACAGAATCCATTATACCCGTGTGCCCTCCCGCCTGCGGCCAGGCGCGGTACGGAGGCGCCGCGGCCGTGGCGCCCTCTGTAGGGACTGTCCTTGCTTTGTTGGGGCTGTACGTGGGAATGGCTTTTAGAAGAATCACCAATACCAAACATTAA
Protein
MDRNIYVYVFYTLLLYFTCNVSAQRTPTISHITQEQIRDIGGQVDLECSVHYAQEFPVVWTKRDRLKTIESIPLSTNAGLIIKDSRFSLRHDEATATFTLSIRDIQENDAGWYQCQVLMSISNKIAAEVELQVRRPPIISDNSTRSIVASEGESVQMECYAGGFPPPKISWRRENNAILPTGGSIYRGNVLKISAVHKEDRGTYYCVADNGVGKGARRNINLEVEFAPVISVPRPRLSQALQYDMDLECHVEAYPPPAITWLKDGIQLSNNQHYRISHFATADEFTDTTIRVITIEKRQYGLFTCKAQNKLGTAEGNVELTESIIPVCPPACGQARYGGAAAVAPSVGTVLALLGLYVGMAFRRITNTKH
Summary
Description
Required for normal tracheal development and maintenance of the trans-epithelial diffusion barrier. Functions as a homophilic cell-adhesion molecule. May play a role in early neuronal differentiation and axon outgrowth.
Keywords
Cell adhesion
Cell membrane
Direct protein sequencing
Disulfide bond
Glycoprotein
GPI-anchor
Immunoglobulin domain
Lipoprotein
Membrane
Repeat
Signal
Complete proteome
Developmental protein
Reference proteome
Feature
chain Lachesin
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
A0A2A4JEZ3
S4PE94
A0A194RP80
A0A194PSG5
A0A1Y1KFY5
A0A212F4V7
+ More
E0VLF8 A0A1Y1KGJ7 D2A4T5 A0A2J7RT88 Q26474 A0A1W4X5H3 V9IKJ5 A0A088AD67 A0A3L8DYV5 A0A336KA52 U5EKE0 A0A067RG08 A0A224XST1 K7J4Y7 A0A1L8E4B6 A0A1S3DEJ8 T1HFD8 A0A232ETM3 E2ABV3 E9IXZ9 A0A2A3ELN1 F4WM47 A0A158P3U6 A0A151XA30 Q16W85 A0A1S4FLW2 A0A310SYQ7 A0A2W1B7C4 E2BCH3 A0A195FU83 A0A1I8JVG0 A0A023EQI4 A0A1B6E1A0 A0A1I8JVS8 A0A1Y9IVE3 A0A2S2QLK8 A0A195BDA2 T1PFT8 A0A2M3Z7D2 A0A2M3Z7P5 A0A0N0U3C9 A0A023ERP4 A0A0A9YFY8 A0A2M4ADA2 A0A2H8TWD4 A0A0L0C2H5 A0A1I8PYN4 W8AXG6 A0A1Y1KAK4 A0A0A1XS42 A0A2M4DM53 A0A1Q3FP08 A0A2W1B9N7 A0A2M4BRC7 A0A034W1E0 B0WNS4 Q7QJG1 A0A0K8UJD3 A0A1J1IV80 A0A0N7ZL50 A0A0N7ZVU9 A0A0P5CX73 B3MEM8 A0A0P4YT49 A0A0N8CKI0 B4MFG8 A0A0P5A0D4 A0A0P5A673 A0A3B0J3T0 A0A0P5PR45 A0A0N8CL30 A0A0P5X3D3 A0A2J7RT94 A0A0P6ABA2 A0A0P6I961 A0A0P5HBU1 A0A0P4Z5Y1 Q28Y34 A0A0K8TRV7 A0A195DCH4 B4MYB0 A0A0C9PP76 A0A0P5UWH1 A0A1B0G0D9 E9GM88 A0A026W5J6 J9K778 A0A0J9RC05 B4P596 B3NS03 Q24372 A0A2S2P7T5 A0A1W4U6M0 B4KR82
E0VLF8 A0A1Y1KGJ7 D2A4T5 A0A2J7RT88 Q26474 A0A1W4X5H3 V9IKJ5 A0A088AD67 A0A3L8DYV5 A0A336KA52 U5EKE0 A0A067RG08 A0A224XST1 K7J4Y7 A0A1L8E4B6 A0A1S3DEJ8 T1HFD8 A0A232ETM3 E2ABV3 E9IXZ9 A0A2A3ELN1 F4WM47 A0A158P3U6 A0A151XA30 Q16W85 A0A1S4FLW2 A0A310SYQ7 A0A2W1B7C4 E2BCH3 A0A195FU83 A0A1I8JVG0 A0A023EQI4 A0A1B6E1A0 A0A1I8JVS8 A0A1Y9IVE3 A0A2S2QLK8 A0A195BDA2 T1PFT8 A0A2M3Z7D2 A0A2M3Z7P5 A0A0N0U3C9 A0A023ERP4 A0A0A9YFY8 A0A2M4ADA2 A0A2H8TWD4 A0A0L0C2H5 A0A1I8PYN4 W8AXG6 A0A1Y1KAK4 A0A0A1XS42 A0A2M4DM53 A0A1Q3FP08 A0A2W1B9N7 A0A2M4BRC7 A0A034W1E0 B0WNS4 Q7QJG1 A0A0K8UJD3 A0A1J1IV80 A0A0N7ZL50 A0A0N7ZVU9 A0A0P5CX73 B3MEM8 A0A0P4YT49 A0A0N8CKI0 B4MFG8 A0A0P5A0D4 A0A0P5A673 A0A3B0J3T0 A0A0P5PR45 A0A0N8CL30 A0A0P5X3D3 A0A2J7RT94 A0A0P6ABA2 A0A0P6I961 A0A0P5HBU1 A0A0P4Z5Y1 Q28Y34 A0A0K8TRV7 A0A195DCH4 B4MYB0 A0A0C9PP76 A0A0P5UWH1 A0A1B0G0D9 E9GM88 A0A026W5J6 J9K778 A0A0J9RC05 B4P596 B3NS03 Q24372 A0A2S2P7T5 A0A1W4U6M0 B4KR82
Pubmed
23622113
26354079
28004739
22118469
20566863
18362917
+ More
19820115 8223276 30249741 24845553 20075255 28648823 20798317 21282665 21719571 21347285 17510324 28756777 24945155 25315136 25401762 26823975 26108605 24495485 25830018 25348373 12364791 17994087 15632085 26369729 21292972 24508170 22936249 17550304 10731132 12537572 12537569 14681183
19820115 8223276 30249741 24845553 20075255 28648823 20798317 21282665 21719571 21347285 17510324 28756777 24945155 25315136 25401762 26823975 26108605 24495485 25830018 25348373 12364791 17994087 15632085 26369729 21292972 24508170 22936249 17550304 10731132 12537572 12537569 14681183
EMBL
NWSH01001831
PCG69972.1
GAIX01003266
JAA89294.1
KQ459875
KPJ19653.1
+ More
KQ459593 KPI96252.1 GEZM01087701 JAV58525.1 AGBW02010301 OWR48771.1 DS235271 EEB14214.1 GEZM01087700 JAV58526.1 KQ971344 EFA05180.1 NEVH01000006 PNF44050.1 L13256 JR049625 AEY61021.1 QOIP01000002 RLU25403.1 UFQS01000164 UFQT01000164 SSX00682.1 SSX21062.1 GANO01001849 JAB58022.1 KK852658 KDR19146.1 GFTR01005397 JAW11029.1 GFDF01000712 JAV13372.1 ACPB03021649 NNAY01002270 OXU21667.1 GL438382 EFN69091.1 GL766836 EFZ14560.1 KZ288220 PBC32182.1 GL888217 EGI64691.1 ADTU01001341 KQ982353 KYQ57214.1 CH477572 EAT38865.1 KQ759773 OAD62917.1 KZ150256 PZC71792.1 GL447283 EFN86599.1 KQ981264 KYN44018.1 GAPW01002235 JAC11363.1 GEDC01017258 GEDC01005603 JAS20040.1 JAS31695.1 GGMS01009385 MBY78588.1 KQ976519 KYM82165.1 KA647682 AFP62311.1 GGFM01003685 MBW24436.1 GGFM01003707 MBW24458.1 KQ435888 KOX69521.1 GAPW01002234 JAC11364.1 GBHO01041170 GBHO01041169 GBHO01013058 GBRD01017715 GBRD01017714 GDHC01018522 JAG02434.1 JAG02435.1 JAG30546.1 JAG48112.1 JAQ00107.1 GGFK01005443 MBW38764.1 GFXV01006047 MBW17852.1 JRES01000975 KNC26558.1 GAMC01012905 JAB93650.1 GEZM01087703 GEZM01087702 JAV58522.1 GBXI01014726 GBXI01000562 JAC99565.1 JAD13730.1 GGFL01014427 MBW78605.1 GFDL01005716 JAV29329.1 KZ150311 PZC71461.1 GGFJ01006478 MBW55619.1 GAKP01009576 JAC49376.1 DS232014 EDS31915.1 AAAB01008807 EAA04652.4 GDHF01025517 JAI26797.1 CVRI01000059 CRL03046.1 GDIP01234737 JAI88664.1 GDIP01207545 JAJ15857.1 GDIP01168452 JAJ54950.1 CH902619 EDV35492.1 GDIP01224796 JAI98605.1 GDIP01122807 GDIP01110009 JAL80907.1 CH940667 EDW57139.1 GDIP01222342 GDIP01218592 GDIP01201893 GDIP01130953 GDIP01105305 GDIP01102459 GDIP01094558 GDIP01084663 GDIP01080909 GDIP01047422 LRGB01000261 JAJ04810.1 KZS19971.1 GDIP01203491 JAJ19911.1 OUUW01000001 SPP73993.1 GDIQ01145689 JAL06037.1 GDIP01121207 JAL82507.1 GDIP01078314 JAM25401.1 PNF44049.1 GDIP01031756 JAM71959.1 GDIQ01024557 JAN70180.1 GDIQ01230100 JAK21625.1 GDIP01219987 JAJ03415.1 CM000071 EAL26132.3 GDAI01000720 JAI16883.1 KQ980989 KYN10591.1 CH963894 EDW77099.2 GBYB01002993 JAG72760.1 GDIP01107851 JAL95863.1 CCAG010016559 GL732552 EFX79321.1 KK107399 EZA51347.1 ABLF02038223 CM002911 KMY93214.1 CM000158 EDW90758.1 CH954179 EDV56305.2 L13255 AE013599 AY051829 BT003177 GGMR01012871 MBY25490.1 CH933808 EDW08269.1
KQ459593 KPI96252.1 GEZM01087701 JAV58525.1 AGBW02010301 OWR48771.1 DS235271 EEB14214.1 GEZM01087700 JAV58526.1 KQ971344 EFA05180.1 NEVH01000006 PNF44050.1 L13256 JR049625 AEY61021.1 QOIP01000002 RLU25403.1 UFQS01000164 UFQT01000164 SSX00682.1 SSX21062.1 GANO01001849 JAB58022.1 KK852658 KDR19146.1 GFTR01005397 JAW11029.1 GFDF01000712 JAV13372.1 ACPB03021649 NNAY01002270 OXU21667.1 GL438382 EFN69091.1 GL766836 EFZ14560.1 KZ288220 PBC32182.1 GL888217 EGI64691.1 ADTU01001341 KQ982353 KYQ57214.1 CH477572 EAT38865.1 KQ759773 OAD62917.1 KZ150256 PZC71792.1 GL447283 EFN86599.1 KQ981264 KYN44018.1 GAPW01002235 JAC11363.1 GEDC01017258 GEDC01005603 JAS20040.1 JAS31695.1 GGMS01009385 MBY78588.1 KQ976519 KYM82165.1 KA647682 AFP62311.1 GGFM01003685 MBW24436.1 GGFM01003707 MBW24458.1 KQ435888 KOX69521.1 GAPW01002234 JAC11364.1 GBHO01041170 GBHO01041169 GBHO01013058 GBRD01017715 GBRD01017714 GDHC01018522 JAG02434.1 JAG02435.1 JAG30546.1 JAG48112.1 JAQ00107.1 GGFK01005443 MBW38764.1 GFXV01006047 MBW17852.1 JRES01000975 KNC26558.1 GAMC01012905 JAB93650.1 GEZM01087703 GEZM01087702 JAV58522.1 GBXI01014726 GBXI01000562 JAC99565.1 JAD13730.1 GGFL01014427 MBW78605.1 GFDL01005716 JAV29329.1 KZ150311 PZC71461.1 GGFJ01006478 MBW55619.1 GAKP01009576 JAC49376.1 DS232014 EDS31915.1 AAAB01008807 EAA04652.4 GDHF01025517 JAI26797.1 CVRI01000059 CRL03046.1 GDIP01234737 JAI88664.1 GDIP01207545 JAJ15857.1 GDIP01168452 JAJ54950.1 CH902619 EDV35492.1 GDIP01224796 JAI98605.1 GDIP01122807 GDIP01110009 JAL80907.1 CH940667 EDW57139.1 GDIP01222342 GDIP01218592 GDIP01201893 GDIP01130953 GDIP01105305 GDIP01102459 GDIP01094558 GDIP01084663 GDIP01080909 GDIP01047422 LRGB01000261 JAJ04810.1 KZS19971.1 GDIP01203491 JAJ19911.1 OUUW01000001 SPP73993.1 GDIQ01145689 JAL06037.1 GDIP01121207 JAL82507.1 GDIP01078314 JAM25401.1 PNF44049.1 GDIP01031756 JAM71959.1 GDIQ01024557 JAN70180.1 GDIQ01230100 JAK21625.1 GDIP01219987 JAJ03415.1 CM000071 EAL26132.3 GDAI01000720 JAI16883.1 KQ980989 KYN10591.1 CH963894 EDW77099.2 GBYB01002993 JAG72760.1 GDIP01107851 JAL95863.1 CCAG010016559 GL732552 EFX79321.1 KK107399 EZA51347.1 ABLF02038223 CM002911 KMY93214.1 CM000158 EDW90758.1 CH954179 EDV56305.2 L13255 AE013599 AY051829 BT003177 GGMR01012871 MBY25490.1 CH933808 EDW08269.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000009046
UP000007266
+ More
UP000235965 UP000192223 UP000005203 UP000279307 UP000027135 UP000002358 UP000079169 UP000015103 UP000215335 UP000000311 UP000242457 UP000007755 UP000005205 UP000075809 UP000008820 UP000008237 UP000078541 UP000075903 UP000076407 UP000075920 UP000078540 UP000095301 UP000053105 UP000037069 UP000095300 UP000002320 UP000007062 UP000183832 UP000007801 UP000008792 UP000076858 UP000268350 UP000001819 UP000078492 UP000007798 UP000092444 UP000000305 UP000053097 UP000007819 UP000002282 UP000008711 UP000000803 UP000192221 UP000009192
UP000235965 UP000192223 UP000005203 UP000279307 UP000027135 UP000002358 UP000079169 UP000015103 UP000215335 UP000000311 UP000242457 UP000007755 UP000005205 UP000075809 UP000008820 UP000008237 UP000078541 UP000075903 UP000076407 UP000075920 UP000078540 UP000095301 UP000053105 UP000037069 UP000095300 UP000002320 UP000007062 UP000183832 UP000007801 UP000008792 UP000076858 UP000268350 UP000001819 UP000078492 UP000007798 UP000092444 UP000000305 UP000053097 UP000007819 UP000002282 UP000008711 UP000000803 UP000192221 UP000009192
PRIDE
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A2A4JEZ3
S4PE94
A0A194RP80
A0A194PSG5
A0A1Y1KFY5
A0A212F4V7
+ More
E0VLF8 A0A1Y1KGJ7 D2A4T5 A0A2J7RT88 Q26474 A0A1W4X5H3 V9IKJ5 A0A088AD67 A0A3L8DYV5 A0A336KA52 U5EKE0 A0A067RG08 A0A224XST1 K7J4Y7 A0A1L8E4B6 A0A1S3DEJ8 T1HFD8 A0A232ETM3 E2ABV3 E9IXZ9 A0A2A3ELN1 F4WM47 A0A158P3U6 A0A151XA30 Q16W85 A0A1S4FLW2 A0A310SYQ7 A0A2W1B7C4 E2BCH3 A0A195FU83 A0A1I8JVG0 A0A023EQI4 A0A1B6E1A0 A0A1I8JVS8 A0A1Y9IVE3 A0A2S2QLK8 A0A195BDA2 T1PFT8 A0A2M3Z7D2 A0A2M3Z7P5 A0A0N0U3C9 A0A023ERP4 A0A0A9YFY8 A0A2M4ADA2 A0A2H8TWD4 A0A0L0C2H5 A0A1I8PYN4 W8AXG6 A0A1Y1KAK4 A0A0A1XS42 A0A2M4DM53 A0A1Q3FP08 A0A2W1B9N7 A0A2M4BRC7 A0A034W1E0 B0WNS4 Q7QJG1 A0A0K8UJD3 A0A1J1IV80 A0A0N7ZL50 A0A0N7ZVU9 A0A0P5CX73 B3MEM8 A0A0P4YT49 A0A0N8CKI0 B4MFG8 A0A0P5A0D4 A0A0P5A673 A0A3B0J3T0 A0A0P5PR45 A0A0N8CL30 A0A0P5X3D3 A0A2J7RT94 A0A0P6ABA2 A0A0P6I961 A0A0P5HBU1 A0A0P4Z5Y1 Q28Y34 A0A0K8TRV7 A0A195DCH4 B4MYB0 A0A0C9PP76 A0A0P5UWH1 A0A1B0G0D9 E9GM88 A0A026W5J6 J9K778 A0A0J9RC05 B4P596 B3NS03 Q24372 A0A2S2P7T5 A0A1W4U6M0 B4KR82
E0VLF8 A0A1Y1KGJ7 D2A4T5 A0A2J7RT88 Q26474 A0A1W4X5H3 V9IKJ5 A0A088AD67 A0A3L8DYV5 A0A336KA52 U5EKE0 A0A067RG08 A0A224XST1 K7J4Y7 A0A1L8E4B6 A0A1S3DEJ8 T1HFD8 A0A232ETM3 E2ABV3 E9IXZ9 A0A2A3ELN1 F4WM47 A0A158P3U6 A0A151XA30 Q16W85 A0A1S4FLW2 A0A310SYQ7 A0A2W1B7C4 E2BCH3 A0A195FU83 A0A1I8JVG0 A0A023EQI4 A0A1B6E1A0 A0A1I8JVS8 A0A1Y9IVE3 A0A2S2QLK8 A0A195BDA2 T1PFT8 A0A2M3Z7D2 A0A2M3Z7P5 A0A0N0U3C9 A0A023ERP4 A0A0A9YFY8 A0A2M4ADA2 A0A2H8TWD4 A0A0L0C2H5 A0A1I8PYN4 W8AXG6 A0A1Y1KAK4 A0A0A1XS42 A0A2M4DM53 A0A1Q3FP08 A0A2W1B9N7 A0A2M4BRC7 A0A034W1E0 B0WNS4 Q7QJG1 A0A0K8UJD3 A0A1J1IV80 A0A0N7ZL50 A0A0N7ZVU9 A0A0P5CX73 B3MEM8 A0A0P4YT49 A0A0N8CKI0 B4MFG8 A0A0P5A0D4 A0A0P5A673 A0A3B0J3T0 A0A0P5PR45 A0A0N8CL30 A0A0P5X3D3 A0A2J7RT94 A0A0P6ABA2 A0A0P6I961 A0A0P5HBU1 A0A0P4Z5Y1 Q28Y34 A0A0K8TRV7 A0A195DCH4 B4MYB0 A0A0C9PP76 A0A0P5UWH1 A0A1B0G0D9 E9GM88 A0A026W5J6 J9K778 A0A0J9RC05 B4P596 B3NS03 Q24372 A0A2S2P7T5 A0A1W4U6M0 B4KR82
PDB
6EG1
E-value=6.1668e-34,
Score=360
Ontologies
GO
Topology
Subcellular location
Cell membrane
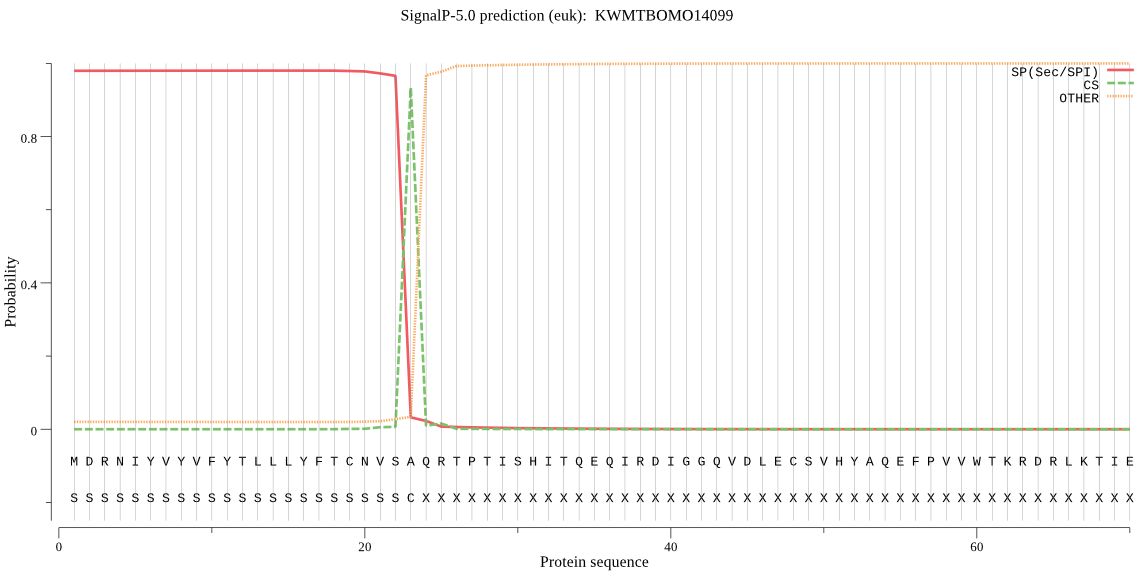
SignalP
Position: 1 - 23,
Likelihood: 0.979972
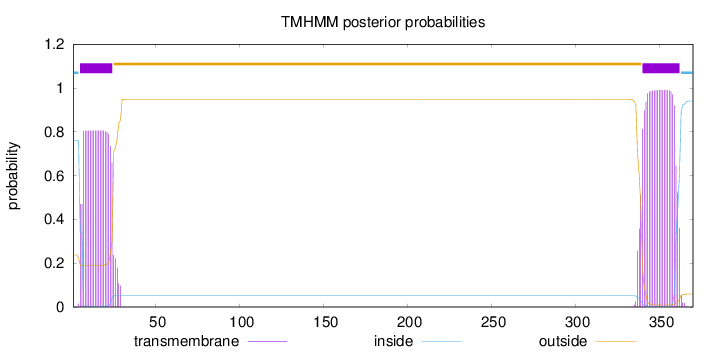
Length:
370
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
38.2137699999999
Exp number, first 60 AAs:
16.05793
Total prob of N-in:
0.76214
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 24
outside
25 - 339
TMhelix
340 - 362
inside
363 - 370
Population Genetic Test Statistics
Pi
270.40823
Theta
187.881939
Tajima's D
1.865534
CLR
0.369461
CSRT
0.857857107144643
Interpretation
Uncertain
