Gene
KWMTBOMO14098 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011209
Annotation
zinc-containing_alcohol_dehydrogenase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.324
Sequence
CDS
ATGGAGGCGGTCGTATTCGATGGAAAAACTTTGACATTAAAATACGATAACAAGTATCCATTGCCGAAGATAGAAAACGACGATGACGTCATTGTGAAAGTGGAGTACTCCGGGATATGTGGGACTGATCTACACATTGTACAGGGTGAGTTTCCGGCTTCCAAAGAGCGCCCCCTACCGCTTGGTCACGAATTCAGTGGCACCATTACGGATGTTGGGAAGAAGTCTGTCTTCAGGAAAGGTCAGAAGGTCGTCGTGGATCCGAACAGAGCCTGCAGCCTCTGTGATTTCTGTCGTAAAGGAAAATACCAGTACTGCTTGACCGCCGGTATTAACAGTACCGTCGGGATCTGGCGGGACGGAGGCTGGGCCCAGTACGTAAAGGTTCCGCAGGATCAGGTCTACCTCCTACCTGACGGCGTGTCCACGGAGCAAGGTGGCCTATGCGAGCCATACTCTTGCGTGGCGCACGGCTACGATCGCGCTTCCCCTCTACTTGTCGGAGAGAAAATCCTCATCGTCGGAGCCGGAATTATTGGTAACCTGTGGGTCACGTCCCTACATCAACTAGGCCACAGAGACGTGACCGTTTCAGAGATGAATAAAGTCAGACTTGAAATCGTCAACAAACTGGAGACTGGATACAGGTTGGTCACTCCCGATGTTCTGGACAAGGAGAAACAGCTGTATGACGTCATCATCGACTGCACAGGCGTCGGTAAAGTGATGGAGATCAGTTTCAATTATCTTCGTCACGGCGGGAAATACGTGCTATTCGGATGCTGTCCACCGACTCATCAAGCTTCGATAAATCCATTCCAAATTTACGACAAGGAACTGACCATCATCGGCGTTAAGATCAACCCGTTCAGCTTCCCTAACGCCTTGGGCTGGCTGAAGGCCATGGGTAATAGATACGTGAATTATGAAAAACTCGGCGTGAAGACGTACGCTCTATCCGAATACGAAAGCGCTCTGAAAGATTTAAGAGCCGGCCTTGTTTCTAAAGCCATGTTCAAAATCAATTAA
Protein
MEAVVFDGKTLTLKYDNKYPLPKIENDDDVIVKVEYSGICGTDLHIVQGEFPASKERPLPLGHEFSGTITDVGKKSVFRKGQKVVVDPNRACSLCDFCRKGKYQYCLTAGINSTVGIWRDGGWAQYVKVPQDQVYLLPDGVSTEQGGLCEPYSCVAHGYDRASPLLVGEKILIVGAGIIGNLWVTSLHQLGHRDVTVSEMNKVRLEIVNKLETGYRLVTPDVLDKEKQLYDVIIDCTGVGKVMEISFNYLRHGGKYVLFGCCPPTHQASINPFQIYDKELTIIGVKINPFSFPNALGWLKAMGNRYVNYEKLGVKTYALSEYESALKDLRAGLVSKAMFKIN
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the zinc-containing alcohol dehydrogenase family.
Uniprot
H9JNV5
Q1HPY0
A0A194PTF2
A0A194RQH8
A0A2H1VEX6
A0A2A4JDD1
+ More
A0A2A4JEV2 A0A2W1BFB4 A0A076FX82 A0A1L8D6P6 A0A212EYS2 A0A0H4TD19 D6WYR6 A0A067RIH2 A0A1Y1LHH5 A0A1Y1MTA2 A0A2J7QB59 A0A2J7QB60 J3JU03 A0A2J7Q3V3 A0A0T6BEC3 A0A195DCF3 E2BCH2 A0A1W4XMI5 A0A1W4XNC9 F4WM48 A0A1W4XCY3 A0A195FUQ0 A0A158P3T5 A0A0L7R9Z1 A0A154PGF5 K7J4Y6 A0A151XA21 A0A232ETH8 A0A310SXA2 A0A2A3EK51 Q4PP82 E9IXZ8 A0A088AD00 A0A2P8YLT1 A0A1B6K2C4 A0A1B6K131 A0A1B6EU91 A0A1B6K2I3 A0A1B6FK96 A0A151IGJ8 A0A1B6M8Y1 T1IDC8 A0A067R903 A0A026W5R6 A0A0P4VVH5 A0A1B6DMR7 A0A0A9ZHE3 A0A146LM48 A0A0A9ZGG8 A0A1B6DVG1 A0A224XFD1 A0A1B6E5Z1 E2ABV2 A0A3M6V5H8 D6WYR5 A0A2R7WIE0 D6WYR7 A0A023FA93 A0A0N0BD22 C3ZND0 A0A1B6JK94 A0A1B6E9I5 A0A161MS66 A0A1B6DI54 A0A3M6V5L2 A0A2B4RBR9 A7SKJ4 A0A195BC75 A7RKY5 V3ZZP2 H2XLJ4 E4XUX5 E4YSY7 V4BPT2 V4AIZ7 C3ZNC9 A0A1B6E8M6 V9IKU3 H2ZFL7 E0VK41 A0A1W2W147 A0A267G8E0 A0A267EJV9 A0A369SK53 B3RSK2 H2ZFL5 A0A2N3WDH6
A0A2A4JEV2 A0A2W1BFB4 A0A076FX82 A0A1L8D6P6 A0A212EYS2 A0A0H4TD19 D6WYR6 A0A067RIH2 A0A1Y1LHH5 A0A1Y1MTA2 A0A2J7QB59 A0A2J7QB60 J3JU03 A0A2J7Q3V3 A0A0T6BEC3 A0A195DCF3 E2BCH2 A0A1W4XMI5 A0A1W4XNC9 F4WM48 A0A1W4XCY3 A0A195FUQ0 A0A158P3T5 A0A0L7R9Z1 A0A154PGF5 K7J4Y6 A0A151XA21 A0A232ETH8 A0A310SXA2 A0A2A3EK51 Q4PP82 E9IXZ8 A0A088AD00 A0A2P8YLT1 A0A1B6K2C4 A0A1B6K131 A0A1B6EU91 A0A1B6K2I3 A0A1B6FK96 A0A151IGJ8 A0A1B6M8Y1 T1IDC8 A0A067R903 A0A026W5R6 A0A0P4VVH5 A0A1B6DMR7 A0A0A9ZHE3 A0A146LM48 A0A0A9ZGG8 A0A1B6DVG1 A0A224XFD1 A0A1B6E5Z1 E2ABV2 A0A3M6V5H8 D6WYR5 A0A2R7WIE0 D6WYR7 A0A023FA93 A0A0N0BD22 C3ZND0 A0A1B6JK94 A0A1B6E9I5 A0A161MS66 A0A1B6DI54 A0A3M6V5L2 A0A2B4RBR9 A7SKJ4 A0A195BC75 A7RKY5 V3ZZP2 H2XLJ4 E4XUX5 E4YSY7 V4BPT2 V4AIZ7 C3ZNC9 A0A1B6E8M6 V9IKU3 H2ZFL7 E0VK41 A0A1W2W147 A0A267G8E0 A0A267EJV9 A0A369SK53 B3RSK2 H2ZFL5 A0A2N3WDH6
Pubmed
19121390
26354079
28756777
24817326
22118469
18362917
+ More
19820115 24845553 28004739 22516182 23537049 20798317 21719571 21347285 20075255 28648823 21282665 29403074 24508170 30249741 27129103 25401762 26823975 30382153 25474469 18563158 17615350 23254933 12481130 15114417 21097902 20566863 30042472 18719581
19820115 24845553 28004739 22516182 23537049 20798317 21719571 21347285 20075255 28648823 21282665 29403074 24508170 30249741 27129103 25401762 26823975 30382153 25474469 18563158 17615350 23254933 12481130 15114417 21097902 20566863 30042472 18719581
EMBL
BABH01009866
BABH01009867
DQ443272
ABF51361.1
KQ459593
KPI96253.1
+ More
KQ459875 KPJ19654.1 ODYU01002199 SOQ39379.1 NWSH01001831 PCG69971.1 PCG69970.1 KZ150311 PZC71460.1 KF960799 AII22001.1 GEYN01000011 JAV02118.1 AGBW02011465 OWR46607.1 KP899556 AKQ06155.1 KQ971357 EFA08453.1 KK852613 KDR20240.1 GEZM01055167 JAV73099.1 GEZM01021882 JAV88841.1 NEVH01016302 PNF25814.1 PNF25813.1 APGK01054661 BT126714 KB741251 AEE61676.1 ENN71848.1 NEVH01018421 PNF23264.1 LJIG01001258 KRT85682.1 KQ980989 KYN10590.1 GL447283 EFN86598.1 GL888217 EGI64692.1 KQ981264 KYN44017.1 ADTU01001340 KQ414620 KOC67685.1 KQ434889 KZC10270.1 KQ982353 KYQ57215.1 NNAY01002270 OXU21668.1 KQ759773 OAD62916.1 KZ288220 PBC32183.1 DQ062231 AAY63986.1 GL766836 EFZ14557.1 PYGN01000503 PSN45209.1 GECU01002128 JAT05579.1 GECU01002550 JAT05157.1 GECZ01028247 JAS41522.1 GECU01002055 JAT05652.1 GECZ01019170 JAS50599.1 KQ977726 KYN00320.1 GEBQ01007584 JAT32393.1 ACPB03008198 KK852618 KDR20140.1 KK107399 QOIP01000002 EZA51348.1 RLU25871.1 GDKW01001842 JAI54753.1 GEDC01010317 GEDC01004818 JAS26981.1 JAS32480.1 GBHO01000411 GBHO01000407 GBRD01003872 GDHC01000553 JAG43193.1 JAG43197.1 JAG61949.1 JAQ18076.1 GDHC01009621 JAQ09008.1 GBHO01000410 GBHO01000408 GBRD01004976 JAG43194.1 JAG43196.1 JAG60845.1 GEDC01007647 JAS29651.1 GFTR01005241 JAW11185.1 GEDC01003989 GEDC01002939 JAS33309.1 JAS34359.1 GL438382 EFN69090.1 RCHS01000065 RMX61233.1 EFA08452.1 KK854878 PTY19414.1 EFA08454.1 GBBI01000387 JAC18325.1 KQ435888 KOX69522.1 GG666650 EEN45958.1 GECU01008068 JAS99638.1 GEDC01002702 JAS34596.1 GEMB01001028 JAS02119.1 GEDC01011935 JAS25363.1 RMX61232.1 LSMT01000685 PFX15101.1 DS469688 EDO35781.1 KQ976519 KYM82166.1 DS469517 EDO47765.1 KB201362 ESO97023.1 FN653193 CBY13522.1 FN655263 CBY38576.1 KB202325 ESO90859.1 ESO97022.1 EEN45957.1 GEDC01003053 JAS34245.1 JR049815 AEY61086.1 DS235239 EEB13746.1 NIVC01000481 PAA82305.1 NIVC01002054 PAA61254.1 NOWV01000009 RDD46413.1 DS985243 EDV26525.1 PJMY01000003 PKV91930.1
KQ459875 KPJ19654.1 ODYU01002199 SOQ39379.1 NWSH01001831 PCG69971.1 PCG69970.1 KZ150311 PZC71460.1 KF960799 AII22001.1 GEYN01000011 JAV02118.1 AGBW02011465 OWR46607.1 KP899556 AKQ06155.1 KQ971357 EFA08453.1 KK852613 KDR20240.1 GEZM01055167 JAV73099.1 GEZM01021882 JAV88841.1 NEVH01016302 PNF25814.1 PNF25813.1 APGK01054661 BT126714 KB741251 AEE61676.1 ENN71848.1 NEVH01018421 PNF23264.1 LJIG01001258 KRT85682.1 KQ980989 KYN10590.1 GL447283 EFN86598.1 GL888217 EGI64692.1 KQ981264 KYN44017.1 ADTU01001340 KQ414620 KOC67685.1 KQ434889 KZC10270.1 KQ982353 KYQ57215.1 NNAY01002270 OXU21668.1 KQ759773 OAD62916.1 KZ288220 PBC32183.1 DQ062231 AAY63986.1 GL766836 EFZ14557.1 PYGN01000503 PSN45209.1 GECU01002128 JAT05579.1 GECU01002550 JAT05157.1 GECZ01028247 JAS41522.1 GECU01002055 JAT05652.1 GECZ01019170 JAS50599.1 KQ977726 KYN00320.1 GEBQ01007584 JAT32393.1 ACPB03008198 KK852618 KDR20140.1 KK107399 QOIP01000002 EZA51348.1 RLU25871.1 GDKW01001842 JAI54753.1 GEDC01010317 GEDC01004818 JAS26981.1 JAS32480.1 GBHO01000411 GBHO01000407 GBRD01003872 GDHC01000553 JAG43193.1 JAG43197.1 JAG61949.1 JAQ18076.1 GDHC01009621 JAQ09008.1 GBHO01000410 GBHO01000408 GBRD01004976 JAG43194.1 JAG43196.1 JAG60845.1 GEDC01007647 JAS29651.1 GFTR01005241 JAW11185.1 GEDC01003989 GEDC01002939 JAS33309.1 JAS34359.1 GL438382 EFN69090.1 RCHS01000065 RMX61233.1 EFA08452.1 KK854878 PTY19414.1 EFA08454.1 GBBI01000387 JAC18325.1 KQ435888 KOX69522.1 GG666650 EEN45958.1 GECU01008068 JAS99638.1 GEDC01002702 JAS34596.1 GEMB01001028 JAS02119.1 GEDC01011935 JAS25363.1 RMX61232.1 LSMT01000685 PFX15101.1 DS469688 EDO35781.1 KQ976519 KYM82166.1 DS469517 EDO47765.1 KB201362 ESO97023.1 FN653193 CBY13522.1 FN655263 CBY38576.1 KB202325 ESO90859.1 ESO97022.1 EEN45957.1 GEDC01003053 JAS34245.1 JR049815 AEY61086.1 DS235239 EEB13746.1 NIVC01000481 PAA82305.1 NIVC01002054 PAA61254.1 NOWV01000009 RDD46413.1 DS985243 EDV26525.1 PJMY01000003 PKV91930.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000007266
+ More
UP000027135 UP000235965 UP000019118 UP000078492 UP000008237 UP000192223 UP000007755 UP000078541 UP000005205 UP000053825 UP000076502 UP000002358 UP000075809 UP000215335 UP000242457 UP000005203 UP000245037 UP000078542 UP000015103 UP000053097 UP000279307 UP000000311 UP000275408 UP000053105 UP000001554 UP000225706 UP000001593 UP000078540 UP000030746 UP000008144 UP000007875 UP000009046 UP000215902 UP000253843 UP000009022 UP000233750
UP000027135 UP000235965 UP000019118 UP000078492 UP000008237 UP000192223 UP000007755 UP000078541 UP000005205 UP000053825 UP000076502 UP000002358 UP000075809 UP000215335 UP000242457 UP000005203 UP000245037 UP000078542 UP000015103 UP000053097 UP000279307 UP000000311 UP000275408 UP000053105 UP000001554 UP000225706 UP000001593 UP000078540 UP000030746 UP000008144 UP000007875 UP000009046 UP000215902 UP000253843 UP000009022 UP000233750
PRIDE
Interpro
ProteinModelPortal
H9JNV5
Q1HPY0
A0A194PTF2
A0A194RQH8
A0A2H1VEX6
A0A2A4JDD1
+ More
A0A2A4JEV2 A0A2W1BFB4 A0A076FX82 A0A1L8D6P6 A0A212EYS2 A0A0H4TD19 D6WYR6 A0A067RIH2 A0A1Y1LHH5 A0A1Y1MTA2 A0A2J7QB59 A0A2J7QB60 J3JU03 A0A2J7Q3V3 A0A0T6BEC3 A0A195DCF3 E2BCH2 A0A1W4XMI5 A0A1W4XNC9 F4WM48 A0A1W4XCY3 A0A195FUQ0 A0A158P3T5 A0A0L7R9Z1 A0A154PGF5 K7J4Y6 A0A151XA21 A0A232ETH8 A0A310SXA2 A0A2A3EK51 Q4PP82 E9IXZ8 A0A088AD00 A0A2P8YLT1 A0A1B6K2C4 A0A1B6K131 A0A1B6EU91 A0A1B6K2I3 A0A1B6FK96 A0A151IGJ8 A0A1B6M8Y1 T1IDC8 A0A067R903 A0A026W5R6 A0A0P4VVH5 A0A1B6DMR7 A0A0A9ZHE3 A0A146LM48 A0A0A9ZGG8 A0A1B6DVG1 A0A224XFD1 A0A1B6E5Z1 E2ABV2 A0A3M6V5H8 D6WYR5 A0A2R7WIE0 D6WYR7 A0A023FA93 A0A0N0BD22 C3ZND0 A0A1B6JK94 A0A1B6E9I5 A0A161MS66 A0A1B6DI54 A0A3M6V5L2 A0A2B4RBR9 A7SKJ4 A0A195BC75 A7RKY5 V3ZZP2 H2XLJ4 E4XUX5 E4YSY7 V4BPT2 V4AIZ7 C3ZNC9 A0A1B6E8M6 V9IKU3 H2ZFL7 E0VK41 A0A1W2W147 A0A267G8E0 A0A267EJV9 A0A369SK53 B3RSK2 H2ZFL5 A0A2N3WDH6
A0A2A4JEV2 A0A2W1BFB4 A0A076FX82 A0A1L8D6P6 A0A212EYS2 A0A0H4TD19 D6WYR6 A0A067RIH2 A0A1Y1LHH5 A0A1Y1MTA2 A0A2J7QB59 A0A2J7QB60 J3JU03 A0A2J7Q3V3 A0A0T6BEC3 A0A195DCF3 E2BCH2 A0A1W4XMI5 A0A1W4XNC9 F4WM48 A0A1W4XCY3 A0A195FUQ0 A0A158P3T5 A0A0L7R9Z1 A0A154PGF5 K7J4Y6 A0A151XA21 A0A232ETH8 A0A310SXA2 A0A2A3EK51 Q4PP82 E9IXZ8 A0A088AD00 A0A2P8YLT1 A0A1B6K2C4 A0A1B6K131 A0A1B6EU91 A0A1B6K2I3 A0A1B6FK96 A0A151IGJ8 A0A1B6M8Y1 T1IDC8 A0A067R903 A0A026W5R6 A0A0P4VVH5 A0A1B6DMR7 A0A0A9ZHE3 A0A146LM48 A0A0A9ZGG8 A0A1B6DVG1 A0A224XFD1 A0A1B6E5Z1 E2ABV2 A0A3M6V5H8 D6WYR5 A0A2R7WIE0 D6WYR7 A0A023FA93 A0A0N0BD22 C3ZND0 A0A1B6JK94 A0A1B6E9I5 A0A161MS66 A0A1B6DI54 A0A3M6V5L2 A0A2B4RBR9 A7SKJ4 A0A195BC75 A7RKY5 V3ZZP2 H2XLJ4 E4XUX5 E4YSY7 V4BPT2 V4AIZ7 C3ZNC9 A0A1B6E8M6 V9IKU3 H2ZFL7 E0VK41 A0A1W2W147 A0A267G8E0 A0A267EJV9 A0A369SK53 B3RSK2 H2ZFL5 A0A2N3WDH6
PDB
4EJM
E-value=2.09368e-39,
Score=407
Ontologies
PANTHER
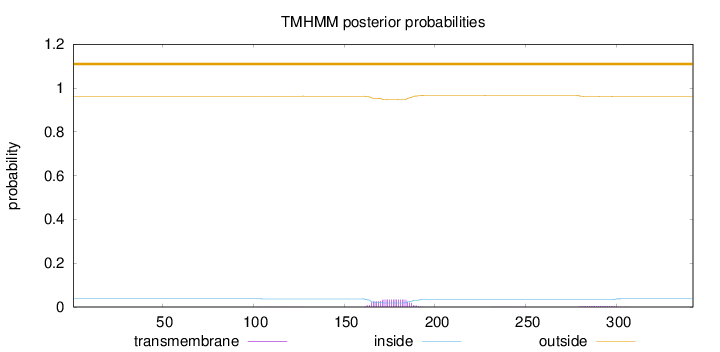
Topology
Length:
342
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.825420000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03757
outside
1 - 342
Population Genetic Test Statistics
Pi
169.31161
Theta
140.962172
Tajima's D
0.653991
CLR
0.101381
CSRT
0.560721963901805
Interpretation
Uncertain
