Gene
KWMTBOMO14094
Pre Gene Modal
BGIBMGA011213
Annotation
PREDICTED:_scavenger_receptor_class_B_member_1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.144
Sequence
CDS
ATGCTGGGATTTATAGCCCTCTCAATAGGATGCTTTATACTGTTCGTGAAGCCATACAACTTTCTGTTTAATCAGAAAGTCCTTCTGCAGGATGGGGGCGAAATCTTCGAGATGTGGAGGAAACCTGAAGTCCAGCTTTATACGAGGGTCTTTCTGTTCAACGTGACTAATTCTGAAGAGTACCTGGCTGGGAAGGATGAGAAAATTAAGCTCCAGGAAGTCGGGCCTTACGTGTACAGAGAGGCTTTGGAACACAAAGTTCAAAGCTTCAACTCGAACGGTACGTTGTCAGCGATTCCCCTGCATCCTCTGACTTGGGTGGAAGAGATGTCCGAGGGCCACAGAGAGGACGACATCGTATATATGCCGCACATAGCTCTACTGAGCATAGCAAACGTAGTCTCCCAGAATTTATTCGCGAGATTCGGCTTAAACAATCTGATTACGTTGACAAACAGCCAGCCCTTGACGAAAATGACGGCCAAAGAGTTCATGATGGGCTATAAGTCCGAGCTGATGACCCTGGGCAACACCTTCCTCCCGAGATGGATTTATTTCGACAAGCTCGGCCTGATCGATCGGATGTATGACTTCGATGGAGACTTCGAGACGGTCTACACAGGCGAAACTGATATCAGACTATCCGGCCTCATCGACACGTACCGCGGCTCCACCGACCTGCCGCAGTGGGAGGGGAAGCACTGCTCAAACGTGCAGTACGCGTCTGACGGCACCAAGTTCAGGGGTGGCGTCACCAAAAACGAAACCTTGCTGTTCTATAGGAAGAGTCTGTGCCGAGCTGCTCCTTTGATCCCTAGCGAGGAGGGCGTCAGAAGCGGCATAAGGGGCGTGAAGTACGTCTTCCCGGAGCACATGCTCGACAACGGCAAACATAACGAAGAGAACAAATGCTTCTGCAGGAATGGAAAATGCCTCCCAGAAGGACTGATCGACGTGGCGGACTGCTACTACGGTTTCCCGATAGCTCTGTCCTATCCCCACTTCTACAAAGGAGACGACGTCCTCTTCAGCAAAGTCGAAGGGCTGACGCCCAAAAAGGAGGAGCATGAGACCAGGCTTTAA
Protein
MLGFIALSIGCFILFVKPYNFLFNQKVLLQDGGEIFEMWRKPEVQLYTRVFLFNVTNSEEYLAGKDEKIKLQEVGPYVYREALEHKVQSFNSNGTLSAIPLHPLTWVEEMSEGHREDDIVYMPHIALLSIANVVSQNLFARFGLNNLITLTNSQPLTKMTAKEFMMGYKSELMTLGNTFLPRWIYFDKLGLIDRMYDFDGDFETVYTGETDIRLSGLIDTYRGSTDLPQWEGKHCSNVQYASDGTKFRGGVTKNETLLFYRKSLCRAAPLIPSEEGVRSGIRGVKYVFPEHMLDNGKHNEENKCFCRNGKCLPEGLIDVADCYYGFPIALSYPHFYKGDDVLFSKVEGLTPKKEEHETRL
Summary
Similarity
Belongs to the CD36 family.
Uniprot
A0A2A4K141
A0A3S2NFK2
A0A194RQ90
A0A194PSH0
A0A2H1WLC8
A0A1J1J3Z6
+ More
A0A139WG73 A0A195FWI4 A0A195EDX3 A0A158NHD6 A0A1B6J672 A0A1B6M9G3 E2B9C5 A0A1B6LBY0 A0A154PEP6 A0A195AZH7 A0A1B6MVM3 A0A1B6IUH7 A0A088AKK6 F4X1X1 A0A2A3EJ06 A0A0L7QQ97 A0A151X8F9 A0A2J7PNU5 A0A0C9QRN9 A0A310SMC7 A0A195D4Q2 A0A0C9QR91 A0A1Y1KLH6 A0A1Y1KJ95 A0A067QYX6 A0A232EZ92 K7IRA8 A0A1Y1KDW8 A0A0M8ZY30 A0A1Y1KDR6 A0A026VWL9 A0A139WFW1 A0A336LYF7 A0A2S2Q087 A0A336L8K1 E0VPT8 A0A1L8DJF3 A0A3L8DH31 A0A182M5B4 A0A182IYY8 A0A1Q3G583 A0A1B6E1D3 A0A1L8DJY4 A0A1L8DJK2 A0A1L8DJL5 B4JQ31 W8APG5 A0A023EWJ5 N6TZF9 W8BDQ7 Q16QP3 A0A1D6T3G5 A0A182LE16 B4MUL6 A0A182XAB2 Q7QCP3 A0A0K8V9K3 A0A0K8W6B6 A0A0A1X9V9 A0A0Q9X4P3 A0A0R1E7T1 A0A182VLS1 A0A182HS57 A0A0M4ECD1 A0A1A9XQQ1 A0A2H8TYJ6 A0A0K8UG95 A0A0A1XA65 A0A2S2PRE0 A0A3B0KNZ1 B4KFF0 A0A0Q9W8H1 A0A182FQX2 W5JE70 A0A0R3NV07 A0A3B0KGS0 J9JX78 B4IT87 A0A0L0C5C4 N6T293 B4LQP2 A0A1A9ZFB7 A0A0J9R641 B4GXB0 B5DK45 A0A0J9R5H2 A0A1A9W5I5 Q8SYC3 A0A182U751 A0A1W4VA08 A0A1B0FR26 A0A0P9ABP3 A0A0A9ZCG5 A0A1I8N7C1
A0A139WG73 A0A195FWI4 A0A195EDX3 A0A158NHD6 A0A1B6J672 A0A1B6M9G3 E2B9C5 A0A1B6LBY0 A0A154PEP6 A0A195AZH7 A0A1B6MVM3 A0A1B6IUH7 A0A088AKK6 F4X1X1 A0A2A3EJ06 A0A0L7QQ97 A0A151X8F9 A0A2J7PNU5 A0A0C9QRN9 A0A310SMC7 A0A195D4Q2 A0A0C9QR91 A0A1Y1KLH6 A0A1Y1KJ95 A0A067QYX6 A0A232EZ92 K7IRA8 A0A1Y1KDW8 A0A0M8ZY30 A0A1Y1KDR6 A0A026VWL9 A0A139WFW1 A0A336LYF7 A0A2S2Q087 A0A336L8K1 E0VPT8 A0A1L8DJF3 A0A3L8DH31 A0A182M5B4 A0A182IYY8 A0A1Q3G583 A0A1B6E1D3 A0A1L8DJY4 A0A1L8DJK2 A0A1L8DJL5 B4JQ31 W8APG5 A0A023EWJ5 N6TZF9 W8BDQ7 Q16QP3 A0A1D6T3G5 A0A182LE16 B4MUL6 A0A182XAB2 Q7QCP3 A0A0K8V9K3 A0A0K8W6B6 A0A0A1X9V9 A0A0Q9X4P3 A0A0R1E7T1 A0A182VLS1 A0A182HS57 A0A0M4ECD1 A0A1A9XQQ1 A0A2H8TYJ6 A0A0K8UG95 A0A0A1XA65 A0A2S2PRE0 A0A3B0KNZ1 B4KFF0 A0A0Q9W8H1 A0A182FQX2 W5JE70 A0A0R3NV07 A0A3B0KGS0 J9JX78 B4IT87 A0A0L0C5C4 N6T293 B4LQP2 A0A1A9ZFB7 A0A0J9R641 B4GXB0 B5DK45 A0A0J9R5H2 A0A1A9W5I5 Q8SYC3 A0A182U751 A0A1W4VA08 A0A1B0FR26 A0A0P9ABP3 A0A0A9ZCG5 A0A1I8N7C1
Pubmed
26354079
18362917
19820115
21347285
20798317
21719571
+ More
28004739 24845553 28648823 20075255 24508170 20566863 30249741 17994087 24495485 24945155 23537049 17510324 20966253 12364791 25830018 17550304 20920257 23761445 15632085 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 26823975 25315136
28004739 24845553 28648823 20075255 24508170 20566863 30249741 17994087 24495485 24945155 23537049 17510324 20966253 12364791 25830018 17550304 20920257 23761445 15632085 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 26823975 25315136
EMBL
NWSH01000254
PCG77991.1
RSAL01000135
RVE46258.1
KQ459875
KPJ19657.1
+ More
KQ459593 KPI96257.1 ODYU01009439 SOQ53869.1 CVRI01000067 CRL06508.1 KQ971348 KYB26889.1 KQ981208 KYN44808.1 KQ979039 KYN23425.1 ADTU01015617 ADTU01015618 ADTU01015619 ADTU01015620 GECU01013046 JAS94660.1 GEBQ01007404 JAT32573.1 GL446493 EFN87706.1 GEBQ01018790 JAT21187.1 KQ434889 KZC10319.1 KQ976694 KYM77643.1 GEBQ01000026 JAT39951.1 GECU01017125 JAS90581.1 GL888555 EGI59558.1 KZ288248 PBC31169.1 KQ414792 KOC60729.1 KQ982409 KYQ56653.1 NEVH01023284 PNF18020.1 GBYB01006384 JAG76151.1 KQ762196 OAD56141.1 KQ976870 KYN07821.1 GBYB01006229 JAG75996.1 GEZM01086191 JAV59687.1 GEZM01086192 JAV59685.1 KK853094 KDR11500.1 NNAY01001554 OXU23617.1 GEZM01086194 JAV59682.1 KQ435827 KOX71919.1 GEZM01086193 JAV59683.1 KK107672 EZA48178.1 KYB26888.1 UFQT01000186 SSX21297.1 GGMS01001943 MBY71146.1 UFQS01002145 UFQT01002145 SSX13321.1 SSX32757.1 DS235379 EEB15394.1 GFDF01007594 JAV06490.1 QOIP01000008 RLU19750.1 AXCM01006435 AXCP01007876 GFDL01000116 JAV34929.1 GEDC01005560 JAS31738.1 GFDF01007424 JAV06660.1 GFDF01007425 JAV06659.1 GFDF01007426 JAV06658.1 CH916372 EDV99011.1 GAMC01018638 GAMC01018637 JAB87917.1 GAPW01000709 JAC12889.1 APGK01055245 KB741261 KB632413 ENN71632.1 ERL95261.1 GAMC01018636 GAMC01018635 JAB87919.1 CH477739 EAT36719.1 APCN01000243 CH963857 EDW76211.2 AAAB01008859 EAA07765.4 GDHF01016841 JAI35473.1 GDHF01005643 JAI46671.1 GBXI01013686 GBXI01006844 JAD00606.1 JAD07448.1 CH933807 KRG03056.1 CH891662 KRK05097.1 CP012523 ALC40414.1 GFXV01007521 MBW19326.1 GDHF01026617 JAI25697.1 GBXI01006724 JAD07568.1 GGMR01019364 MBY31983.1 OUUW01000010 SPP85558.1 EDW12050.1 CH940649 KRF81112.1 ADMH02001643 ETN61638.1 CH379061 KRT04955.1 SPP85559.1 ABLF02026897 ABLF02026902 EDW99477.1 JRES01000973 KNC26639.1 ENN71633.1 ERL95262.1 EDW63426.1 CM002910 KMY91451.1 CH479195 EDW27221.1 EDY70671.1 KMY91452.1 AY071642 AE014134 AAL49264.1 EAA46015.1 CCAG010000954 CH902638 KPU75614.1 GBHO01040299 GBHO01037707 GBHO01002028 GBHO01002023 GDHC01001230 JAG03305.1 JAG05897.1 JAG41576.1 JAG41581.1 JAQ17399.1
KQ459593 KPI96257.1 ODYU01009439 SOQ53869.1 CVRI01000067 CRL06508.1 KQ971348 KYB26889.1 KQ981208 KYN44808.1 KQ979039 KYN23425.1 ADTU01015617 ADTU01015618 ADTU01015619 ADTU01015620 GECU01013046 JAS94660.1 GEBQ01007404 JAT32573.1 GL446493 EFN87706.1 GEBQ01018790 JAT21187.1 KQ434889 KZC10319.1 KQ976694 KYM77643.1 GEBQ01000026 JAT39951.1 GECU01017125 JAS90581.1 GL888555 EGI59558.1 KZ288248 PBC31169.1 KQ414792 KOC60729.1 KQ982409 KYQ56653.1 NEVH01023284 PNF18020.1 GBYB01006384 JAG76151.1 KQ762196 OAD56141.1 KQ976870 KYN07821.1 GBYB01006229 JAG75996.1 GEZM01086191 JAV59687.1 GEZM01086192 JAV59685.1 KK853094 KDR11500.1 NNAY01001554 OXU23617.1 GEZM01086194 JAV59682.1 KQ435827 KOX71919.1 GEZM01086193 JAV59683.1 KK107672 EZA48178.1 KYB26888.1 UFQT01000186 SSX21297.1 GGMS01001943 MBY71146.1 UFQS01002145 UFQT01002145 SSX13321.1 SSX32757.1 DS235379 EEB15394.1 GFDF01007594 JAV06490.1 QOIP01000008 RLU19750.1 AXCM01006435 AXCP01007876 GFDL01000116 JAV34929.1 GEDC01005560 JAS31738.1 GFDF01007424 JAV06660.1 GFDF01007425 JAV06659.1 GFDF01007426 JAV06658.1 CH916372 EDV99011.1 GAMC01018638 GAMC01018637 JAB87917.1 GAPW01000709 JAC12889.1 APGK01055245 KB741261 KB632413 ENN71632.1 ERL95261.1 GAMC01018636 GAMC01018635 JAB87919.1 CH477739 EAT36719.1 APCN01000243 CH963857 EDW76211.2 AAAB01008859 EAA07765.4 GDHF01016841 JAI35473.1 GDHF01005643 JAI46671.1 GBXI01013686 GBXI01006844 JAD00606.1 JAD07448.1 CH933807 KRG03056.1 CH891662 KRK05097.1 CP012523 ALC40414.1 GFXV01007521 MBW19326.1 GDHF01026617 JAI25697.1 GBXI01006724 JAD07568.1 GGMR01019364 MBY31983.1 OUUW01000010 SPP85558.1 EDW12050.1 CH940649 KRF81112.1 ADMH02001643 ETN61638.1 CH379061 KRT04955.1 SPP85559.1 ABLF02026897 ABLF02026902 EDW99477.1 JRES01000973 KNC26639.1 ENN71633.1 ERL95262.1 EDW63426.1 CM002910 KMY91451.1 CH479195 EDW27221.1 EDY70671.1 KMY91452.1 AY071642 AE014134 AAL49264.1 EAA46015.1 CCAG010000954 CH902638 KPU75614.1 GBHO01040299 GBHO01037707 GBHO01002028 GBHO01002023 GDHC01001230 JAG03305.1 JAG05897.1 JAG41576.1 JAG41581.1 JAQ17399.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000183832
UP000007266
+ More
UP000078541 UP000078492 UP000005205 UP000008237 UP000076502 UP000078540 UP000005203 UP000007755 UP000242457 UP000053825 UP000075809 UP000235965 UP000078542 UP000027135 UP000215335 UP000002358 UP000053105 UP000053097 UP000009046 UP000279307 UP000075883 UP000075880 UP000001070 UP000019118 UP000030742 UP000008820 UP000075840 UP000075882 UP000007798 UP000076407 UP000007062 UP000009192 UP000002282 UP000075903 UP000092553 UP000092443 UP000268350 UP000008792 UP000069272 UP000000673 UP000001819 UP000007819 UP000037069 UP000092445 UP000008744 UP000091820 UP000000803 UP000075902 UP000192221 UP000092444 UP000007801 UP000095301
UP000078541 UP000078492 UP000005205 UP000008237 UP000076502 UP000078540 UP000005203 UP000007755 UP000242457 UP000053825 UP000075809 UP000235965 UP000078542 UP000027135 UP000215335 UP000002358 UP000053105 UP000053097 UP000009046 UP000279307 UP000075883 UP000075880 UP000001070 UP000019118 UP000030742 UP000008820 UP000075840 UP000075882 UP000007798 UP000076407 UP000007062 UP000009192 UP000002282 UP000075903 UP000092553 UP000092443 UP000268350 UP000008792 UP000069272 UP000000673 UP000001819 UP000007819 UP000037069 UP000092445 UP000008744 UP000091820 UP000000803 UP000075902 UP000192221 UP000092444 UP000007801 UP000095301
PRIDE
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
A0A2A4K141
A0A3S2NFK2
A0A194RQ90
A0A194PSH0
A0A2H1WLC8
A0A1J1J3Z6
+ More
A0A139WG73 A0A195FWI4 A0A195EDX3 A0A158NHD6 A0A1B6J672 A0A1B6M9G3 E2B9C5 A0A1B6LBY0 A0A154PEP6 A0A195AZH7 A0A1B6MVM3 A0A1B6IUH7 A0A088AKK6 F4X1X1 A0A2A3EJ06 A0A0L7QQ97 A0A151X8F9 A0A2J7PNU5 A0A0C9QRN9 A0A310SMC7 A0A195D4Q2 A0A0C9QR91 A0A1Y1KLH6 A0A1Y1KJ95 A0A067QYX6 A0A232EZ92 K7IRA8 A0A1Y1KDW8 A0A0M8ZY30 A0A1Y1KDR6 A0A026VWL9 A0A139WFW1 A0A336LYF7 A0A2S2Q087 A0A336L8K1 E0VPT8 A0A1L8DJF3 A0A3L8DH31 A0A182M5B4 A0A182IYY8 A0A1Q3G583 A0A1B6E1D3 A0A1L8DJY4 A0A1L8DJK2 A0A1L8DJL5 B4JQ31 W8APG5 A0A023EWJ5 N6TZF9 W8BDQ7 Q16QP3 A0A1D6T3G5 A0A182LE16 B4MUL6 A0A182XAB2 Q7QCP3 A0A0K8V9K3 A0A0K8W6B6 A0A0A1X9V9 A0A0Q9X4P3 A0A0R1E7T1 A0A182VLS1 A0A182HS57 A0A0M4ECD1 A0A1A9XQQ1 A0A2H8TYJ6 A0A0K8UG95 A0A0A1XA65 A0A2S2PRE0 A0A3B0KNZ1 B4KFF0 A0A0Q9W8H1 A0A182FQX2 W5JE70 A0A0R3NV07 A0A3B0KGS0 J9JX78 B4IT87 A0A0L0C5C4 N6T293 B4LQP2 A0A1A9ZFB7 A0A0J9R641 B4GXB0 B5DK45 A0A0J9R5H2 A0A1A9W5I5 Q8SYC3 A0A182U751 A0A1W4VA08 A0A1B0FR26 A0A0P9ABP3 A0A0A9ZCG5 A0A1I8N7C1
A0A139WG73 A0A195FWI4 A0A195EDX3 A0A158NHD6 A0A1B6J672 A0A1B6M9G3 E2B9C5 A0A1B6LBY0 A0A154PEP6 A0A195AZH7 A0A1B6MVM3 A0A1B6IUH7 A0A088AKK6 F4X1X1 A0A2A3EJ06 A0A0L7QQ97 A0A151X8F9 A0A2J7PNU5 A0A0C9QRN9 A0A310SMC7 A0A195D4Q2 A0A0C9QR91 A0A1Y1KLH6 A0A1Y1KJ95 A0A067QYX6 A0A232EZ92 K7IRA8 A0A1Y1KDW8 A0A0M8ZY30 A0A1Y1KDR6 A0A026VWL9 A0A139WFW1 A0A336LYF7 A0A2S2Q087 A0A336L8K1 E0VPT8 A0A1L8DJF3 A0A3L8DH31 A0A182M5B4 A0A182IYY8 A0A1Q3G583 A0A1B6E1D3 A0A1L8DJY4 A0A1L8DJK2 A0A1L8DJL5 B4JQ31 W8APG5 A0A023EWJ5 N6TZF9 W8BDQ7 Q16QP3 A0A1D6T3G5 A0A182LE16 B4MUL6 A0A182XAB2 Q7QCP3 A0A0K8V9K3 A0A0K8W6B6 A0A0A1X9V9 A0A0Q9X4P3 A0A0R1E7T1 A0A182VLS1 A0A182HS57 A0A0M4ECD1 A0A1A9XQQ1 A0A2H8TYJ6 A0A0K8UG95 A0A0A1XA65 A0A2S2PRE0 A0A3B0KNZ1 B4KFF0 A0A0Q9W8H1 A0A182FQX2 W5JE70 A0A0R3NV07 A0A3B0KGS0 J9JX78 B4IT87 A0A0L0C5C4 N6T293 B4LQP2 A0A1A9ZFB7 A0A0J9R641 B4GXB0 B5DK45 A0A0J9R5H2 A0A1A9W5I5 Q8SYC3 A0A182U751 A0A1W4VA08 A0A1B0FR26 A0A0P9ABP3 A0A0A9ZCG5 A0A1I8N7C1
PDB
5UPH
E-value=9.00538e-41,
Score=419
Ontologies
GO
PANTHER
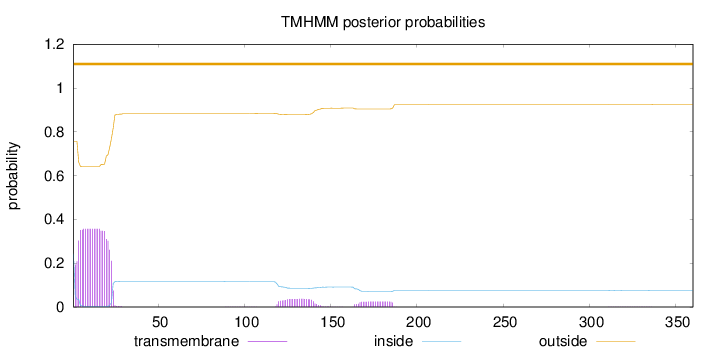
Topology
Length:
360
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.52977
Exp number, first 60 AAs:
7.20203
Total prob of N-in:
0.24346
outside
1 - 360
Population Genetic Test Statistics
Pi
185.236722
Theta
158.863567
Tajima's D
0.509387
CLR
1.180677
CSRT
0.511124443777811
Interpretation
Uncertain
