Gene
KWMTBOMO14088
Pre Gene Modal
BGIBMGA014599
Annotation
PREDICTED:_esterase_FE4-like_isoform_X1_[Bombyx_mori]
Full name
Carboxylic ester hydrolase
Location in the cell
Cytoplasmic Reliability : 1.565
Sequence
CDS
ATGTCATGCATTGTGCAAGTTGAGCAGGGCTTATTGGAGGGAACGAAGCGTCAGAGCTGCCGTGGTCAGTATTACTACAGCTTTGAGGGAATACCGTATGCGAAGCCACCTATTGGAAAGCTGAGATTTAGTAATCCACAAGAACCCGATCAATGGAAAGGAATACGTAGCGCAAAAAGTCCCTCAAACAAATGCGCCCAATTAAATCCTTATTCTAACTCAGCGTTAGTAGGTTCGGAAGACTGTTTGTACTTAAACATTTACACACCTAGTTTGCCAGCTGAAAAACTCGAGAAATTACCAGTAATCTTCTTCGTCCACGGCGGTCGTTTGATTGTAGGCTACGGAGATTATTATACACCGGATTATTTGATTAAAAACGATGTTATTTTAGTAACGTTAAATTACAGGTTAAATGTGTTAGGATTTTTGTGTTTGAATTTACCGGAAGCACCTGGTAACGTTGGAATTAAGGACACCGTCTACGCATTACGTTGGGTCGGTAGAAATATTTCTCGTTTTAATGGCAACCCTGGTAATATAACGGTTTCCGGTGAAAGCGCCGGTGGGGCAATTGTATCTTCGTACTTAACCTCAAAGATGACTGTTGGTCTTTTTAGTAAGGTCATTGCGCAGTCCGGGAATTCAATATCAGATTTGTATTTGATTAACGAGGATCCTATTGAGAAAGCGATAAAATTGGCGTCAGTTTTAGGCAAGGATTTGAAAGATCAACGGTCTTTATACGAGTTCTTAGTACAGGTTCCGTTAGTAGATTTGGTGAATGCGTTCACCCAGCTAGAACTGAGTCGGCCGCCTAGTATTATCAATGCGTATTTCTTACCGGTCGTCGAGAAAAAGTTCGATAACGTCGAAAGTTTTTTCGAGGAACATCCCATAGTGTCCATTAAGAATAATAGAGTACAGAAAGTGCCCGTAATAACAGGTTATACCACTTACGAATCAGCGCTGTTCGTCCAAAAAGACTACTCCGGTAGGATCGTATATGTAGATGACTTTCATAATTTTATCCCTCGATTCTTGGGTGTCGAAAAGAACACGGAAAGATCTAGAGTTTTCCAGAAGAAACTACGCGAATTCTATTTTGCGGGCCGGGAATTGAATGACGATTTGAAGATTGATTATTTGGTAATGATCACGGACGTGTATTTCGTTAGGGATATCATTTATTTCGCTGAATTACTATCAAAACACAATCCTGAGGTGTATCTATATGAATTCGGTTTTGACGGCAACCTGAATACGAGGGTTATGAAGAGTTTGGGAGTGAAGGGAGCGTCGCACGGAGACATCGTGCAGTATCAGTTTTACAGGAAGAGCAAAGCAGAAAAATGCGGGGAGGCTGACGAGAGAACGATTGACTTCTTGAGCGAATGCTGGAGTAATTTTGCTAAATACGGCGTACCAACATGGAGGAATCAGAAAACAAAATGGCTGCCTTACTCAACGAACAATAGACGGACGCTAGTTATAAATGACGAGATAGAATTAAAGAGAGACGTTAATTTGAATCGCATTAAGTTCTGGTACGATATGATCAAGGATAAGAGTAAATTGTGA
Protein
MSCIVQVEQGLLEGTKRQSCRGQYYYSFEGIPYAKPPIGKLRFSNPQEPDQWKGIRSAKSPSNKCAQLNPYSNSALVGSEDCLYLNIYTPSLPAEKLEKLPVIFFVHGGRLIVGYGDYYTPDYLIKNDVILVTLNYRLNVLGFLCLNLPEAPGNVGIKDTVYALRWVGRNISRFNGNPGNITVSGESAGGAIVSSYLTSKMTVGLFSKVIAQSGNSISDLYLINEDPIEKAIKLASVLGKDLKDQRSLYEFLVQVPLVDLVNAFTQLELSRPPSIINAYFLPVVEKKFDNVESFFEEHPIVSIKNNRVQKVPVITGYTTYESALFVQKDYSGRIVYVDDFHNFIPRFLGVEKNTERSRVFQKKLREFYFAGRELNDDLKIDYLVMITDVYFVRDIIYFAELLSKHNPEVYLYEFGFDGNLNTRVMKSLGVKGASHGDIVQYQFYRKSKAEKCGEADERTIDFLSECWSNFAKYGVPTWRNQKTKWLPYSTNNRRTLVINDEIELKRDVNLNRIKFWYDMIKDKSKL
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Feature
chain Carboxylic ester hydrolase
Uniprot
H9JYH9
A0A291P0S9
A0A291P0Z4
A0A2W1B642
A0A2W1BCZ0
A0A2H1V8U9
+ More
A0A194RP89 A0A0B4RXW5 A0A0K0XRV9 A0A0K8TV77 A0A1L8D6S5 A0A1U9X1V5 A0A0L7LQY5 A0A1L8D6R3 A0A194PYN5 A0A1U9X1X8 A0A0C5C1R4 A0A0L7L781 A0A0K8TUN9 A0A291P112 A0A291P0U0 H9J594 D5G3F4 A0A2H1X0W5 A0A1J0M180 A0A2S0D942 A0A291P0R0 D8UUL2 A0A0K0XRL5 A0A2W1BUX0 F8V329 D5G3F1 A0A2A4JW69 A0A1L8D6L5 A0A2A4JXU8 A0A194PYQ2 D5G3F0 A0A0K0XRV7 A0A1U9X1W7 D5G3E6 A0A2S0D938 A0A2A4JYH8 A0A2W1B1E5 A0A2A4JX20 H9J593 A0A1L8D6K5 A0A2U3T8L1 A0A194RF34 B5AK62 B5AK61 A0A2J7RMN6 D2KWC5 D2KWC4 Q1HPP2 A0A2H1V8S9 A0A194Q3V9 A0A0F7QEA8 A0A2J7RMN7 B6D7H0 A0A194RDD6 H9JRG2 A0A0K8TVA5 A0A1J0M120 A0A2J7RMV5 A0A1U9X1X6 G1C2I5 Q59HJ2 A0A2J7RMP3 A0A2I6RJN9 A0A2J7RMU4 A0A194PTN5 A9LS22 A0A0K0XRL4 H9JN84 A0A291P117 A0A3S2NFG0 D3GDM2 A0A076FT12 A0A3S2MAW7 A0A291P0V7 A0A2H1WNU4 A0A194R2U2 A0A0K8TVV4 G3CM70 A0A067RMU8 A0A2A4JIZ5 A0A291P123 A0A2A4J5S2 A0A0K8TUL7 A0A0Y0BA98 A0A0K1YWC6 A0A2A4J274 A0A194R4Z8 A0A109QN31 A0A2J7RMS4 A0A1J0M1C9 Q1W2E3 A0A2H1V381 A0A291P0Y3 A0A2H1VCU4
A0A194RP89 A0A0B4RXW5 A0A0K0XRV9 A0A0K8TV77 A0A1L8D6S5 A0A1U9X1V5 A0A0L7LQY5 A0A1L8D6R3 A0A194PYN5 A0A1U9X1X8 A0A0C5C1R4 A0A0L7L781 A0A0K8TUN9 A0A291P112 A0A291P0U0 H9J594 D5G3F4 A0A2H1X0W5 A0A1J0M180 A0A2S0D942 A0A291P0R0 D8UUL2 A0A0K0XRL5 A0A2W1BUX0 F8V329 D5G3F1 A0A2A4JW69 A0A1L8D6L5 A0A2A4JXU8 A0A194PYQ2 D5G3F0 A0A0K0XRV7 A0A1U9X1W7 D5G3E6 A0A2S0D938 A0A2A4JYH8 A0A2W1B1E5 A0A2A4JX20 H9J593 A0A1L8D6K5 A0A2U3T8L1 A0A194RF34 B5AK62 B5AK61 A0A2J7RMN6 D2KWC5 D2KWC4 Q1HPP2 A0A2H1V8S9 A0A194Q3V9 A0A0F7QEA8 A0A2J7RMN7 B6D7H0 A0A194RDD6 H9JRG2 A0A0K8TVA5 A0A1J0M120 A0A2J7RMV5 A0A1U9X1X6 G1C2I5 Q59HJ2 A0A2J7RMP3 A0A2I6RJN9 A0A2J7RMU4 A0A194PTN5 A9LS22 A0A0K0XRL4 H9JN84 A0A291P117 A0A3S2NFG0 D3GDM2 A0A076FT12 A0A3S2MAW7 A0A291P0V7 A0A2H1WNU4 A0A194R2U2 A0A0K8TVV4 G3CM70 A0A067RMU8 A0A2A4JIZ5 A0A291P123 A0A2A4J5S2 A0A0K8TUL7 A0A0Y0BA98 A0A0K1YWC6 A0A2A4J274 A0A194R4Z8 A0A109QN31 A0A2J7RMS4 A0A1J0M1C9 Q1W2E3 A0A2H1V381 A0A291P0Y3 A0A2H1VCU4
EC Number
3.1.1.-
Pubmed
EMBL
BABH01043679
MF687566
ATJ44492.1
MF687636
ATJ44562.1
KZ150466
+ More
PZC70781.1 PZC70780.1 ODYU01001263 SOQ37239.1 KQ459875 KPJ19663.1 KJ023213 AIY69041.1 KP938876 AKS40357.1 GCVX01000042 JAI18188.1 GEYN01000034 JAV02095.1 KY021847 AQY62736.1 JTDY01000293 KOB77862.1 GEYN01000044 JAV02085.1 KQ459593 KPI96265.1 KY021874 AQY62763.1 KP001159 AJN91204.1 JTDY01002474 KOB71338.1 GCVX01000033 JAI18197.1 MF687632 ATJ44558.1 MF687553 ATJ44479.1 BABH01039690 BABH01039691 FJ997318 ADF43483.1 ODYU01012558 SOQ58953.1 KU375439 APD13881.1 KX015867 ARM65396.1 MF687557 ATJ44483.1 GQ180944 ACT53736.1 KP938878 AKS40359.1 KZ149909 PZC78161.1 JN019808 AEJ18172.1 FJ997315 ADF43480.1 NWSH01000459 PCG76251.1 GEYN01000033 JAV02096.1 PCG76250.1 KQ459586 KPI98158.1 FJ997314 ADF43479.1 KP938875 AKS40356.1 KY021869 AQY62758.1 FJ997310 ADF43475.1 KX015868 ARM65397.1 NWSH01000416 PCG76543.1 PZC70779.1 PCG76541.1 GEYN01000043 JAV02086.1 KX015866 ARM65395.1 KQ460323 KPJ15910.1 EU854155 EU854157 EU854158 ACF98321.1 ACF98323.1 ACF98324.1 EU854154 EU854156 ACF98320.1 ACF98322.1 NEVH01002552 PNF42095.1 AB497080 AB497081 BAI66486.1 BAI66487.1 AB497079 BAI66485.1 DQ443360 ABF51449.1 SOQ37238.1 KPI98095.1 LC017762 BAR64776.1 PNF42088.1 EU854151 ACI16653.1 KPJ15853.1 BABH01016384 GCVX01000034 JAI18196.1 KU375431 APD13873.1 PNF42152.1 KY021878 AQY62767.1 JF728804 AEJ38206.1 AB208651 BAD91555.1 PNF42100.1 KY304476 AUN86407.1 PNF42151.1 KQ459592 KPI96776.1 EU255790 ABX46627.1 KP938873 AKS40354.1 BABH01033219 MF687633 ATJ44559.1 RSAL01000139 RVE46156.1 FJ652454 ACV60238.1 KF960782 AII21984.1 RSAL01000002 RVE55036.1 RVE55089.1 MF687564 ATJ44490.1 ODYU01009893 SOQ54612.1 KQ460845 KPJ12017.1 GCVX01000035 JAI18195.1 HQ122616 AEL33699.1 KK852415 KDR24383.1 NWSH01001228 PCG72037.1 MF687624 ATJ44550.1 NWSH01003029 PCG67086.1 GCVX01000041 JAI18189.1 KU215371 AMB19665.1 KP859354 AKZ17664.1 NWSH01003954 PCG65624.1 KQ460779 KPJ12315.1 KU215373 AMB19667.1 PNF42128.1 KU375427 APD13869.1 DQ445461 ABE01157.2 ODYU01000269 SOQ34744.1 MF687626 ATJ44552.1 ODYU01001848 SOQ38621.1
PZC70781.1 PZC70780.1 ODYU01001263 SOQ37239.1 KQ459875 KPJ19663.1 KJ023213 AIY69041.1 KP938876 AKS40357.1 GCVX01000042 JAI18188.1 GEYN01000034 JAV02095.1 KY021847 AQY62736.1 JTDY01000293 KOB77862.1 GEYN01000044 JAV02085.1 KQ459593 KPI96265.1 KY021874 AQY62763.1 KP001159 AJN91204.1 JTDY01002474 KOB71338.1 GCVX01000033 JAI18197.1 MF687632 ATJ44558.1 MF687553 ATJ44479.1 BABH01039690 BABH01039691 FJ997318 ADF43483.1 ODYU01012558 SOQ58953.1 KU375439 APD13881.1 KX015867 ARM65396.1 MF687557 ATJ44483.1 GQ180944 ACT53736.1 KP938878 AKS40359.1 KZ149909 PZC78161.1 JN019808 AEJ18172.1 FJ997315 ADF43480.1 NWSH01000459 PCG76251.1 GEYN01000033 JAV02096.1 PCG76250.1 KQ459586 KPI98158.1 FJ997314 ADF43479.1 KP938875 AKS40356.1 KY021869 AQY62758.1 FJ997310 ADF43475.1 KX015868 ARM65397.1 NWSH01000416 PCG76543.1 PZC70779.1 PCG76541.1 GEYN01000043 JAV02086.1 KX015866 ARM65395.1 KQ460323 KPJ15910.1 EU854155 EU854157 EU854158 ACF98321.1 ACF98323.1 ACF98324.1 EU854154 EU854156 ACF98320.1 ACF98322.1 NEVH01002552 PNF42095.1 AB497080 AB497081 BAI66486.1 BAI66487.1 AB497079 BAI66485.1 DQ443360 ABF51449.1 SOQ37238.1 KPI98095.1 LC017762 BAR64776.1 PNF42088.1 EU854151 ACI16653.1 KPJ15853.1 BABH01016384 GCVX01000034 JAI18196.1 KU375431 APD13873.1 PNF42152.1 KY021878 AQY62767.1 JF728804 AEJ38206.1 AB208651 BAD91555.1 PNF42100.1 KY304476 AUN86407.1 PNF42151.1 KQ459592 KPI96776.1 EU255790 ABX46627.1 KP938873 AKS40354.1 BABH01033219 MF687633 ATJ44559.1 RSAL01000139 RVE46156.1 FJ652454 ACV60238.1 KF960782 AII21984.1 RSAL01000002 RVE55036.1 RVE55089.1 MF687564 ATJ44490.1 ODYU01009893 SOQ54612.1 KQ460845 KPJ12017.1 GCVX01000035 JAI18195.1 HQ122616 AEL33699.1 KK852415 KDR24383.1 NWSH01001228 PCG72037.1 MF687624 ATJ44550.1 NWSH01003029 PCG67086.1 GCVX01000041 JAI18189.1 KU215371 AMB19665.1 KP859354 AKZ17664.1 NWSH01003954 PCG65624.1 KQ460779 KPJ12315.1 KU215373 AMB19667.1 PNF42128.1 KU375427 APD13869.1 DQ445461 ABE01157.2 ODYU01000269 SOQ34744.1 MF687626 ATJ44552.1 ODYU01001848 SOQ38621.1
Proteomes
Pfam
PF00135 COesterase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JYH9
A0A291P0S9
A0A291P0Z4
A0A2W1B642
A0A2W1BCZ0
A0A2H1V8U9
+ More
A0A194RP89 A0A0B4RXW5 A0A0K0XRV9 A0A0K8TV77 A0A1L8D6S5 A0A1U9X1V5 A0A0L7LQY5 A0A1L8D6R3 A0A194PYN5 A0A1U9X1X8 A0A0C5C1R4 A0A0L7L781 A0A0K8TUN9 A0A291P112 A0A291P0U0 H9J594 D5G3F4 A0A2H1X0W5 A0A1J0M180 A0A2S0D942 A0A291P0R0 D8UUL2 A0A0K0XRL5 A0A2W1BUX0 F8V329 D5G3F1 A0A2A4JW69 A0A1L8D6L5 A0A2A4JXU8 A0A194PYQ2 D5G3F0 A0A0K0XRV7 A0A1U9X1W7 D5G3E6 A0A2S0D938 A0A2A4JYH8 A0A2W1B1E5 A0A2A4JX20 H9J593 A0A1L8D6K5 A0A2U3T8L1 A0A194RF34 B5AK62 B5AK61 A0A2J7RMN6 D2KWC5 D2KWC4 Q1HPP2 A0A2H1V8S9 A0A194Q3V9 A0A0F7QEA8 A0A2J7RMN7 B6D7H0 A0A194RDD6 H9JRG2 A0A0K8TVA5 A0A1J0M120 A0A2J7RMV5 A0A1U9X1X6 G1C2I5 Q59HJ2 A0A2J7RMP3 A0A2I6RJN9 A0A2J7RMU4 A0A194PTN5 A9LS22 A0A0K0XRL4 H9JN84 A0A291P117 A0A3S2NFG0 D3GDM2 A0A076FT12 A0A3S2MAW7 A0A291P0V7 A0A2H1WNU4 A0A194R2U2 A0A0K8TVV4 G3CM70 A0A067RMU8 A0A2A4JIZ5 A0A291P123 A0A2A4J5S2 A0A0K8TUL7 A0A0Y0BA98 A0A0K1YWC6 A0A2A4J274 A0A194R4Z8 A0A109QN31 A0A2J7RMS4 A0A1J0M1C9 Q1W2E3 A0A2H1V381 A0A291P0Y3 A0A2H1VCU4
A0A194RP89 A0A0B4RXW5 A0A0K0XRV9 A0A0K8TV77 A0A1L8D6S5 A0A1U9X1V5 A0A0L7LQY5 A0A1L8D6R3 A0A194PYN5 A0A1U9X1X8 A0A0C5C1R4 A0A0L7L781 A0A0K8TUN9 A0A291P112 A0A291P0U0 H9J594 D5G3F4 A0A2H1X0W5 A0A1J0M180 A0A2S0D942 A0A291P0R0 D8UUL2 A0A0K0XRL5 A0A2W1BUX0 F8V329 D5G3F1 A0A2A4JW69 A0A1L8D6L5 A0A2A4JXU8 A0A194PYQ2 D5G3F0 A0A0K0XRV7 A0A1U9X1W7 D5G3E6 A0A2S0D938 A0A2A4JYH8 A0A2W1B1E5 A0A2A4JX20 H9J593 A0A1L8D6K5 A0A2U3T8L1 A0A194RF34 B5AK62 B5AK61 A0A2J7RMN6 D2KWC5 D2KWC4 Q1HPP2 A0A2H1V8S9 A0A194Q3V9 A0A0F7QEA8 A0A2J7RMN7 B6D7H0 A0A194RDD6 H9JRG2 A0A0K8TVA5 A0A1J0M120 A0A2J7RMV5 A0A1U9X1X6 G1C2I5 Q59HJ2 A0A2J7RMP3 A0A2I6RJN9 A0A2J7RMU4 A0A194PTN5 A9LS22 A0A0K0XRL4 H9JN84 A0A291P117 A0A3S2NFG0 D3GDM2 A0A076FT12 A0A3S2MAW7 A0A291P0V7 A0A2H1WNU4 A0A194R2U2 A0A0K8TVV4 G3CM70 A0A067RMU8 A0A2A4JIZ5 A0A291P123 A0A2A4J5S2 A0A0K8TUL7 A0A0Y0BA98 A0A0K1YWC6 A0A2A4J274 A0A194R4Z8 A0A109QN31 A0A2J7RMS4 A0A1J0M1C9 Q1W2E3 A0A2H1V381 A0A291P0Y3 A0A2H1VCU4
PDB
5TYM
E-value=5.02955e-72,
Score=690
Ontologies
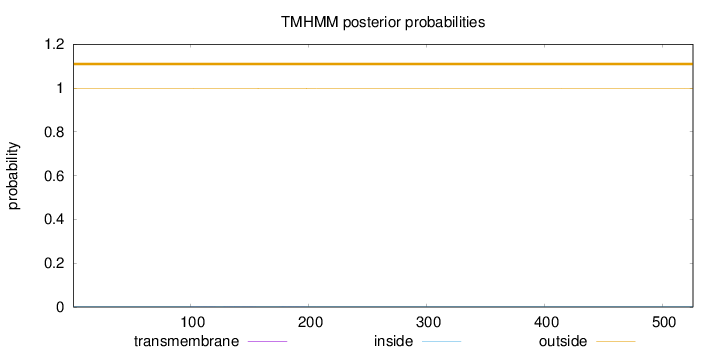
Topology
Length:
526
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02709
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00354
outside
1 - 526
Population Genetic Test Statistics
Pi
291.064823
Theta
180.08661
Tajima's D
2.726866
CLR
0.519305
CSRT
0.96385180740963
Interpretation
Uncertain
