Pre Gene Modal
BGIBMGA011217
Annotation
fatty-acyl_CoA_reductase_1_[Ostrinia_nubilalis]
Full name
Fatty acyl-CoA reductase
Location in the cell
Mitochondrial Reliability : 1.161 PlasmaMembrane Reliability : 1.146
Sequence
CDS
ATGCGGCCTAAGAAATCCGTGAGCCCGGACAAAAGACTGGCTCAATTGTATTCTACTCCTTGCTTCGATAGGCTTCGCAAAGAAAAACCTGGCGTGTTTCAGTCCAAAGTCCTCGTTGTGCCTGGAGATGTAATGGAAGATGGATTAGGTTTATCAGACGAAGACAGGAAACTTCTCGCCGAACGAGTGAATATTGTCTTTCATATAGCTGCTAGTGTGAGATTCGATGACCCCTTGAAGTACGCGGTCAGATTAAACTTGAACGGGACGAAGGAAGTCGTGGAACTAGCTAAGAGTATGCACAACCTATCTTCATTCGTTCACGTGTCGACTAGCTACTCAAACACCAACCGCGAGGTCATCGAGGAGGTGATGTATCCCCCTCACGCTGACTGGAGGGACACCCTGGAGATCTGCGAGAAACTTGACGAGCACACGCTGAGGGTTATGACTCCAAAATATCTCGGAGAGTTGCCCAATACGTACACCTTCAGCAAGCAGTTAGCTGAACACGCTGTATATGAGCAGAGAGGACAACTGCCTGCTGTTATAGTTAGACCGTCTATAGTAATAGCGAGCGCTGATGAACCAATTAGCGGTTGGATTGAGAATTTCAATGGCCCCGTTGGAATATTTGTCGCTTGCGGAAAAGGCATAATGCGTACGTTGTATACGAAGCCAGATATTGTATCTGATTACATACCGGTGGATATATGCGCGAAGAACATTATAGTCGCTGCCTGGGCTAGAGGAACGAAGCCTCTAGAAAGCTCGGACGACGTTGAGATATACAATTGCTGTTCTGGTGAGCTGAACCAAATTAATATCCAGGACCTGATCGAGATGGCCAAGGAGGTAGGCGCCAGTGTCCCCCTGAATGACGCCTTGTGGACAATTGGAGGAAGCATCACCACCTCGGAAAAACTATACTACATCAGGGTATTGGTCCAACATCTCTTGCCGGCGATTTTTGTTGACATAGTCCTTCGTATATTCGGCAAAAAGCCTATGCTCTTCAAAATTCAAAGAAGAATCTACATAGCGAATCTAGCGCTTCGGTATTATATAACGAAGCAATGGACCTTCGAAAATAAAAACTTCCTCGCGCTCAGATCTAAATTACTAGAGCAGGATAAAAAACAATTCTATTACCTACTCGAGGGTAGAGATAAAAAGGATTATTTCTTGAAATGCTGCATCGGTGGAAGGAGGTATCTGATGAAAGAGAGAGACGAAGACATCCCTAAAGCGAAGCTCCGATTTCAGAGAATGATGGTCCTCGACAAGACAGTGCAAGTAATATTCTATGGATTACTCCTATGGTGGTCCATCAAAATATTTAATTATTATTTCGGACTCTACTGTAATTTTTTTGTAGCCTGA
Protein
MRPKKSVSPDKRLAQLYSTPCFDRLRKEKPGVFQSKVLVVPGDVMEDGLGLSDEDRKLLAERVNIVFHIAASVRFDDPLKYAVRLNLNGTKEVVELAKSMHNLSSFVHVSTSYSNTNREVIEEVMYPPHADWRDTLEICEKLDEHTLRVMTPKYLGELPNTYTFSKQLAEHAVYEQRGQLPAVIVRPSIVIASADEPISGWIENFNGPVGIFVACGKGIMRTLYTKPDIVSDYIPVDICAKNIIVAAWARGTKPLESSDDVEIYNCCSGELNQINIQDLIEMAKEVGASVPLNDALWTIGGSITTSEKLYYIRVLVQHLLPAIFVDIVLRIFGKKPMLFKIQRRIYIANLALRYYITKQWTFENKNFLALRSKLLEQDKKQFYYLLEGRDKKDYFLKCCIGGRRYLMKERDEDIPKAKLRFQRMMVLDKTVQVIFYGLLLWWSIKIFNYYFGLYCNFFVA
Summary
Description
Catalyzes the reduction of fatty acyl-CoA to fatty alcohols.
Catalytic Activity
a long-chain fatty acyl-CoA + 2 H(+) + 2 NADPH = a long-chain primary fatty alcohol + CoA + 2 NADP(+)
Similarity
Belongs to the fatty acyl-CoA reductase family.
Uniprot
A0A3S2NWB0
D9D5H4
A0A2A4K2X0
A0A291P0T9
A0A291P0W8
A0A194PTF8
+ More
A0A1L8D6J1 A0A291P0S7 A0A2A4K2J5 A0A194RP84 A0A2H1W7P9 A0A2H1VIF0 A0A212F0B3 A0A1W4WCD3 D6WYQ7 A0A182YH19 A0A182W5X8 A0A182RVQ2 A0A2W1BB54 A0A182FJU1 A0A182QWR0 A0A1S4F214 A0A1Q3FZP3 A0A182T8Q8 A0A1Q3FZV2 A0A182HAJ7 Q7Q2V5 A0A182MZT1 A0A182H739 A0A1B0D476 E2ANI0 A0A084VR72 A0A2J7RI68 A0A182PWI9 A0A067QZV4 B0WYE5 A0A232EY48 A0A2J7RI66 A0A151X3W7 Q17I01 A0A182M7X1 A0A1B6CCS7 A0A3L8DK87 A0A0A9Y3V1 A0A336M1N3 A0A193CHN5 A0A2P8Y5S4 A0A0L7RAZ7 E2BYG6 X1WJJ1 A0A336MN30 B4MEM2 F4WQP0 A0A1B6DEA6 A0A2J7RI77 A0A1B6EWI8 B4L1G7 A0A0L0CAW3 A0A0M4FAK5 D9MWM8 A0A1I8P540 T1PK20 A0A1I8NAX1 A0A088AU73 A0A1W4UW92 K7IMK8 A0A023F0J0 A0A2S2Q9S1 A0A026WLT8 A0A034WLE4 B3MY80 A0A3B0J6S2 W8C494 B4H0C0 Q29H47 A0A0J7K250 A0A158NRZ6 B4JM90 A0A2J7PLY4 A0A1J1HS41 T1I5B7 A0A224XJQ7 A0A0C9PTE7 B4Q1E3 A0A1B6IBF2 A0A336KKV2 B4MTB8 A0A310S7P0 W5JL84 B0WB95 B0WB93 Q17AH3 A0A1I8PJD5 Q17AH5 A0A2M4BL52
A0A1L8D6J1 A0A291P0S7 A0A2A4K2J5 A0A194RP84 A0A2H1W7P9 A0A2H1VIF0 A0A212F0B3 A0A1W4WCD3 D6WYQ7 A0A182YH19 A0A182W5X8 A0A182RVQ2 A0A2W1BB54 A0A182FJU1 A0A182QWR0 A0A1S4F214 A0A1Q3FZP3 A0A182T8Q8 A0A1Q3FZV2 A0A182HAJ7 Q7Q2V5 A0A182MZT1 A0A182H739 A0A1B0D476 E2ANI0 A0A084VR72 A0A2J7RI68 A0A182PWI9 A0A067QZV4 B0WYE5 A0A232EY48 A0A2J7RI66 A0A151X3W7 Q17I01 A0A182M7X1 A0A1B6CCS7 A0A3L8DK87 A0A0A9Y3V1 A0A336M1N3 A0A193CHN5 A0A2P8Y5S4 A0A0L7RAZ7 E2BYG6 X1WJJ1 A0A336MN30 B4MEM2 F4WQP0 A0A1B6DEA6 A0A2J7RI77 A0A1B6EWI8 B4L1G7 A0A0L0CAW3 A0A0M4FAK5 D9MWM8 A0A1I8P540 T1PK20 A0A1I8NAX1 A0A088AU73 A0A1W4UW92 K7IMK8 A0A023F0J0 A0A2S2Q9S1 A0A026WLT8 A0A034WLE4 B3MY80 A0A3B0J6S2 W8C494 B4H0C0 Q29H47 A0A0J7K250 A0A158NRZ6 B4JM90 A0A2J7PLY4 A0A1J1HS41 T1I5B7 A0A224XJQ7 A0A0C9PTE7 B4Q1E3 A0A1B6IBF2 A0A336KKV2 B4MTB8 A0A310S7P0 W5JL84 B0WB95 B0WB93 Q17AH3 A0A1I8PJD5 Q17AH5 A0A2M4BL52
EC Number
1.2.1.84
Pubmed
28986331
26354079
22118469
18362917
19820115
25244985
+ More
28756777 17510324 26483478 12364791 14747013 17210077 20798317 24438588 24845553 28648823 30249741 25401762 27271970 29403074 17994087 21719571 26108605 20542116 25315136 20075255 25474469 24508170 25348373 24495485 15632085 21347285 17550304 20920257 23761445
28756777 17510324 26483478 12364791 14747013 17210077 20798317 24438588 24845553 28648823 30249741 25401762 27271970 29403074 17994087 21719571 26108605 20542116 25315136 20075255 25474469 24508170 25348373 24495485 15632085 21347285 17550304 20920257 23761445
EMBL
RSAL01000135
RVE46259.1
GU808250
ADI82774.1
NWSH01000254
PCG77992.1
+ More
MF687544 ATJ44470.1 MF687600 ATJ44526.1 KQ459593 KPI96258.1 GEYN01000142 JAV01987.1 MF687543 ATJ44469.1 PCG77993.1 KQ459875 KPJ19658.1 ODYU01006856 SOQ49078.1 ODYU01002489 SOQ40034.1 AGBW02011124 OWR47189.1 KQ971357 EFA08449.1 KZ150519 PZC70617.1 AXCN02001406 GFDL01002010 JAV33035.1 GFDL01002009 JAV33036.1 JXUM01122828 KQ566648 KXJ69945.1 AAAB01008967 EAA13114.5 JXUM01115484 JXUM01115485 KQ565885 KXJ70568.1 AJVK01024160 GL441227 EFN65009.1 ATLV01015505 KE525018 KFB40466.1 NEVH01003505 PNF40529.1 KK852809 KDR15980.1 DS232187 EDS37022.1 NNAY01001677 OXU23253.1 PNF40528.1 KQ982562 KYQ54969.1 CH477244 EAT46275.1 AXCM01002370 GEDC01026034 JAS11264.1 QOIP01000007 RLU20840.1 GBHO01016745 GBRD01003868 JAG26859.1 JAG61953.1 UFQS01000395 UFQT01000395 SSX03595.1 SSX23960.1 KU963290 ANN23960.1 PYGN01000896 PSN39593.1 KQ414617 KOC68039.1 GL451478 EFN79250.1 ABLF02038949 ABLF02038951 ABLF02038964 ABLF02057284 UFQT01001371 SSX30233.1 CH940664 EDW62997.1 GL888275 EGI63403.1 GEDC01013326 JAS23972.1 PNF40526.1 GECZ01027461 JAS42308.1 CH933810 EDW06688.2 JRES01000755 KNC28614.1 CP012528 ALC49445.1 HM483392 ADJ56409.1 KA648435 AFP63064.1 GBBI01003720 JAC14992.1 GGMS01005285 MBY74488.1 KK107152 EZA57010.1 GAKP01002566 JAC56386.1 CH902632 EDV32574.1 OUUW01000003 SPP77737.1 GAMC01005674 JAC00882.1 CH479200 EDW29715.1 CH379064 EAL31911.1 LBMM01016171 KMQ84518.1 ADTU01024364 ADTU01024365 ADTU01024366 CH916371 EDV91851.1 NEVH01024421 PNF17348.1 CVRI01000020 CRK90871.1 ACPB03003261 GFTR01006368 JAW10058.1 GBYB01004642 GBYB01004643 JAG74409.1 JAG74410.1 CM000162 EDX02432.1 GECU01023474 JAS84232.1 UFQS01000581 UFQT01000581 SSX05119.1 SSX25480.1 CH963851 EDW75357.2 KQ776822 OAD52159.1 ADMH02001118 ETN64063.1 DS231878 EDS42239.1 EDS42237.1 CH477335 EAT43249.1 EAT43247.1 GGFJ01004658 MBW53799.1
MF687544 ATJ44470.1 MF687600 ATJ44526.1 KQ459593 KPI96258.1 GEYN01000142 JAV01987.1 MF687543 ATJ44469.1 PCG77993.1 KQ459875 KPJ19658.1 ODYU01006856 SOQ49078.1 ODYU01002489 SOQ40034.1 AGBW02011124 OWR47189.1 KQ971357 EFA08449.1 KZ150519 PZC70617.1 AXCN02001406 GFDL01002010 JAV33035.1 GFDL01002009 JAV33036.1 JXUM01122828 KQ566648 KXJ69945.1 AAAB01008967 EAA13114.5 JXUM01115484 JXUM01115485 KQ565885 KXJ70568.1 AJVK01024160 GL441227 EFN65009.1 ATLV01015505 KE525018 KFB40466.1 NEVH01003505 PNF40529.1 KK852809 KDR15980.1 DS232187 EDS37022.1 NNAY01001677 OXU23253.1 PNF40528.1 KQ982562 KYQ54969.1 CH477244 EAT46275.1 AXCM01002370 GEDC01026034 JAS11264.1 QOIP01000007 RLU20840.1 GBHO01016745 GBRD01003868 JAG26859.1 JAG61953.1 UFQS01000395 UFQT01000395 SSX03595.1 SSX23960.1 KU963290 ANN23960.1 PYGN01000896 PSN39593.1 KQ414617 KOC68039.1 GL451478 EFN79250.1 ABLF02038949 ABLF02038951 ABLF02038964 ABLF02057284 UFQT01001371 SSX30233.1 CH940664 EDW62997.1 GL888275 EGI63403.1 GEDC01013326 JAS23972.1 PNF40526.1 GECZ01027461 JAS42308.1 CH933810 EDW06688.2 JRES01000755 KNC28614.1 CP012528 ALC49445.1 HM483392 ADJ56409.1 KA648435 AFP63064.1 GBBI01003720 JAC14992.1 GGMS01005285 MBY74488.1 KK107152 EZA57010.1 GAKP01002566 JAC56386.1 CH902632 EDV32574.1 OUUW01000003 SPP77737.1 GAMC01005674 JAC00882.1 CH479200 EDW29715.1 CH379064 EAL31911.1 LBMM01016171 KMQ84518.1 ADTU01024364 ADTU01024365 ADTU01024366 CH916371 EDV91851.1 NEVH01024421 PNF17348.1 CVRI01000020 CRK90871.1 ACPB03003261 GFTR01006368 JAW10058.1 GBYB01004642 GBYB01004643 JAG74409.1 JAG74410.1 CM000162 EDX02432.1 GECU01023474 JAS84232.1 UFQS01000581 UFQT01000581 SSX05119.1 SSX25480.1 CH963851 EDW75357.2 KQ776822 OAD52159.1 ADMH02001118 ETN64063.1 DS231878 EDS42239.1 EDS42237.1 CH477335 EAT43249.1 EAT43247.1 GGFJ01004658 MBW53799.1
Proteomes
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
UP000192223
+ More
UP000007266 UP000076408 UP000075920 UP000075900 UP000069272 UP000075886 UP000075901 UP000069940 UP000249989 UP000007062 UP000075884 UP000092462 UP000000311 UP000030765 UP000235965 UP000075885 UP000027135 UP000002320 UP000215335 UP000075809 UP000008820 UP000075883 UP000279307 UP000245037 UP000053825 UP000008237 UP000007819 UP000008792 UP000007755 UP000009192 UP000037069 UP000092553 UP000095300 UP000095301 UP000005203 UP000192221 UP000002358 UP000053097 UP000007801 UP000268350 UP000008744 UP000001819 UP000036403 UP000005205 UP000001070 UP000183832 UP000015103 UP000002282 UP000007798 UP000000673
UP000007266 UP000076408 UP000075920 UP000075900 UP000069272 UP000075886 UP000075901 UP000069940 UP000249989 UP000007062 UP000075884 UP000092462 UP000000311 UP000030765 UP000235965 UP000075885 UP000027135 UP000002320 UP000215335 UP000075809 UP000008820 UP000075883 UP000279307 UP000245037 UP000053825 UP000008237 UP000007819 UP000008792 UP000007755 UP000009192 UP000037069 UP000092553 UP000095300 UP000095301 UP000005203 UP000192221 UP000002358 UP000053097 UP000007801 UP000268350 UP000008744 UP000001819 UP000036403 UP000005205 UP000001070 UP000183832 UP000015103 UP000002282 UP000007798 UP000000673
Pfam
Interpro
IPR036291
NAD(P)-bd_dom_sf
+ More
IPR013120 Male_sterile_NAD-bd
IPR033640 FAR_C
IPR026055 FAR
IPR036870 Ribosomal_S18_sf
IPR001648 Ribosomal_S18
IPR003903 UIM_dom
IPR009075 AcylCo_DH/oxidase_C
IPR036465 vWFA_dom_sf
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR013786 AcylCoA_DH/ox_N
IPR006089 Acyl-CoA_DH_CS
IPR002035 VWF_A
IPR002110 Ankyrin_rpt
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR013120 Male_sterile_NAD-bd
IPR033640 FAR_C
IPR026055 FAR
IPR036870 Ribosomal_S18_sf
IPR001648 Ribosomal_S18
IPR003903 UIM_dom
IPR009075 AcylCo_DH/oxidase_C
IPR036465 vWFA_dom_sf
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR013786 AcylCoA_DH/ox_N
IPR006089 Acyl-CoA_DH_CS
IPR002035 VWF_A
IPR002110 Ankyrin_rpt
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
SUPFAM
Gene 3D
ProteinModelPortal
A0A3S2NWB0
D9D5H4
A0A2A4K2X0
A0A291P0T9
A0A291P0W8
A0A194PTF8
+ More
A0A1L8D6J1 A0A291P0S7 A0A2A4K2J5 A0A194RP84 A0A2H1W7P9 A0A2H1VIF0 A0A212F0B3 A0A1W4WCD3 D6WYQ7 A0A182YH19 A0A182W5X8 A0A182RVQ2 A0A2W1BB54 A0A182FJU1 A0A182QWR0 A0A1S4F214 A0A1Q3FZP3 A0A182T8Q8 A0A1Q3FZV2 A0A182HAJ7 Q7Q2V5 A0A182MZT1 A0A182H739 A0A1B0D476 E2ANI0 A0A084VR72 A0A2J7RI68 A0A182PWI9 A0A067QZV4 B0WYE5 A0A232EY48 A0A2J7RI66 A0A151X3W7 Q17I01 A0A182M7X1 A0A1B6CCS7 A0A3L8DK87 A0A0A9Y3V1 A0A336M1N3 A0A193CHN5 A0A2P8Y5S4 A0A0L7RAZ7 E2BYG6 X1WJJ1 A0A336MN30 B4MEM2 F4WQP0 A0A1B6DEA6 A0A2J7RI77 A0A1B6EWI8 B4L1G7 A0A0L0CAW3 A0A0M4FAK5 D9MWM8 A0A1I8P540 T1PK20 A0A1I8NAX1 A0A088AU73 A0A1W4UW92 K7IMK8 A0A023F0J0 A0A2S2Q9S1 A0A026WLT8 A0A034WLE4 B3MY80 A0A3B0J6S2 W8C494 B4H0C0 Q29H47 A0A0J7K250 A0A158NRZ6 B4JM90 A0A2J7PLY4 A0A1J1HS41 T1I5B7 A0A224XJQ7 A0A0C9PTE7 B4Q1E3 A0A1B6IBF2 A0A336KKV2 B4MTB8 A0A310S7P0 W5JL84 B0WB95 B0WB93 Q17AH3 A0A1I8PJD5 Q17AH5 A0A2M4BL52
A0A1L8D6J1 A0A291P0S7 A0A2A4K2J5 A0A194RP84 A0A2H1W7P9 A0A2H1VIF0 A0A212F0B3 A0A1W4WCD3 D6WYQ7 A0A182YH19 A0A182W5X8 A0A182RVQ2 A0A2W1BB54 A0A182FJU1 A0A182QWR0 A0A1S4F214 A0A1Q3FZP3 A0A182T8Q8 A0A1Q3FZV2 A0A182HAJ7 Q7Q2V5 A0A182MZT1 A0A182H739 A0A1B0D476 E2ANI0 A0A084VR72 A0A2J7RI68 A0A182PWI9 A0A067QZV4 B0WYE5 A0A232EY48 A0A2J7RI66 A0A151X3W7 Q17I01 A0A182M7X1 A0A1B6CCS7 A0A3L8DK87 A0A0A9Y3V1 A0A336M1N3 A0A193CHN5 A0A2P8Y5S4 A0A0L7RAZ7 E2BYG6 X1WJJ1 A0A336MN30 B4MEM2 F4WQP0 A0A1B6DEA6 A0A2J7RI77 A0A1B6EWI8 B4L1G7 A0A0L0CAW3 A0A0M4FAK5 D9MWM8 A0A1I8P540 T1PK20 A0A1I8NAX1 A0A088AU73 A0A1W4UW92 K7IMK8 A0A023F0J0 A0A2S2Q9S1 A0A026WLT8 A0A034WLE4 B3MY80 A0A3B0J6S2 W8C494 B4H0C0 Q29H47 A0A0J7K250 A0A158NRZ6 B4JM90 A0A2J7PLY4 A0A1J1HS41 T1I5B7 A0A224XJQ7 A0A0C9PTE7 B4Q1E3 A0A1B6IBF2 A0A336KKV2 B4MTB8 A0A310S7P0 W5JL84 B0WB95 B0WB93 Q17AH3 A0A1I8PJD5 Q17AH5 A0A2M4BL52
PDB
4W4T
E-value=0.001627,
Score=99
Ontologies
GO
PANTHER
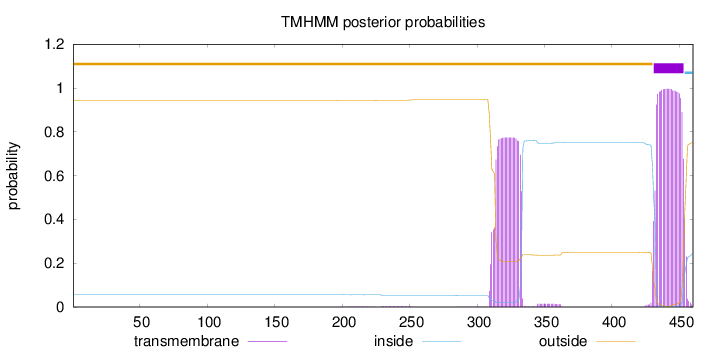
Topology
Length:
460
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
38.83979
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.05650
outside
1 - 430
TMhelix
431 - 453
inside
454 - 460
Population Genetic Test Statistics
Pi
152.337565
Theta
139.287121
Tajima's D
0.628739
CLR
0.414322
CSRT
0.556522173891305
Interpretation
Uncertain
