Gene
KWMTBOMO14080 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011218
Annotation
PREDICTED:_alpha-esterase_48_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.433
Sequence
CDS
ATGATATCGGCCGTGAACGAATTCCTAGATGATCTTCGTGGTGGCAGGATGTCCGAGTCGCCTCTAGTCACGGTGGAGCAAGGGCAGCTCCAGGGACGGATCGTAAACAGCCCGTCCGGGAAGGCCTTCTACAGTTTCCAAGGAATACCATATGCGAAACCCCCATTGGGGTCTTTGCGATTTAAGGCACCTAAACCAGCGGAAACTTGGGAAGGAATCCGAGATGCCACAGCAGAGGGAAACATCGCTCCGCAAATAGACACATCGCTCACGAAAACATATACTGGCGATGAAAACTGCCTATATTTAAACGTATATACCCCACATATCGCCGGAAACCTACCCGTAATGATTTGGATCCATGGTGGGGCCTTCAAATGGGGCTCAGGCAATGAAACCCTATATGGCCCCGATTATCTGGTTGAGAAAGACGTTGTCGTTGTTACCCTCAACTACAGATGTGGACCTTTGGGTTTCTTATGCCTGAATACCCCTGAAGTGCCAGGAAACGCTGGACTCAAAGACGTGGTCCAAGCACTGAGGTGGCTAAAACAGAATATAAAGAGTTTCGGTGGAGATCCTGATAATTTCACTGTGTTCGGTCAAAGCGCCGGTGGGGCCATTGTAACGATACTAACTGCTAGTCCACTGTCTAAAAACATGATTAATAAGGCTATTGTACAATCCGGTACCGGTATATCAAAATGGGCTGTACAAAATGAGCCATTGACCTGTGCCAAGGCTTTGGCAAGTCATTTAGGTTGCGAAGCTGACAACGTTGACGAAGTTCTAGAGTTCTTGAACACAGTCACGGCTAAAGAACTTGTCGAGGCAACGGAAATCGTTAATTCCTTCGATAGTTTAGTTGTCGATCAGAATAACTTTTTCTCTGTTGTCGTCGAGAAAGAATTCCCGGGTGTCGAAGCTGTACTGACGGAGCCTCTTTTAGATTTCTTAACGTCAGGACGTACAGCCGAAATTCCTATAATGATCGGCTCGACAACGCTCGAACTTCTAACTAACCTAAGACCGAGTGATTTGCAAATGTTTATCCCTTCAGATTTGAATATTGAAAAGGATTCAGACGAGTCCTTAGCCATTGCTGAAAATTTAAAAGGCTTATATTTCACAAGCGATACAGAAGAAAACAAAGCCGAAGGTCTAAACAGGCTCCATTCTGATTTATTACTGAATATAAACATCAATAGATACATCAAATATTTAGTTCAAAATTCAAATCAGCCAATATATTTTTATAAATTTGATTACGTTGGCCAATTCAACTTCGCCCATAAGTCATTTTTTGATACAGAACTAAAATACGCCCTGCACATGGACGATTTAGGGTATTTGTTCAAAAATGACTTCCAGAAAGACGTGGATGACCCAAGCCCTCAAGATGTTAAGATGAGGGAAAGAATGGTCAGACTTTGGACTAATTTCGCCAAATTTGGAAATCCCACCCCAGAAGAAAACCACTATCTCCCAACGAAATGGCTACCGGTTACCAATGATACTCTCTACTACCTAAACTTGGGTCAAGAGCTGAATCTTCTCCAAAACCCTGACGAGGAGATAATGAAATTCTGGGAAGATTTATACAGTAAACACTTCAAAATTTGGGAGCATACTAAAATAAATACCATCGAGCCACCCAGGAAACTTGTCTATCCTGATAACGAAGCTACTGAAGAACAAGCTTCCATTGATGACGGACAACCTAGCCTTGAAGAAAAAGTCGTTTTTGAAGAAATAGTGCCTATCAGTTCGGTACTAAACCCACCAGTTAACGGTGATGCTAAATCGAACGCAGATTTTAAGCCGAAGATTTCGAATGAAATTAAAATGGCTCAAACTTCTGCACCTAAAGACGTCGTGAGAGCCAGCGACCCGCCCGAAGACGACCTACCAAAGAACATAGGGGCCCCACAACCTCCAGAACCCTGGGACGGCATCAGGGATGCCACGGCGGAAGGGAACGTGTGTGCTCAAATCGATCCGGTCTTCGCGAAATCCTATGTCGGCGATGAAAACTGCTTGTTCCTAAATGTGTACACCCCTAGCACTGATGGAGCATTCCTCCCGGTCATGATTTGGATCCACGGTGGTGGGTTCAAGTGGGGATCCGGCAATACCAACTTGTATGGACCAGATTTTCTAGTTGACAGAGATGTCGTGGTGGTGACTATCAATTATAGATGTGGTGCTTTAGGGTTCTTAAGTCTGAACACGCCTGAAGTCCCAGGGAATGCTGGGATCAAAGATATAGTTCAAGCAATTAGATGGGTGAAAGATAATATCCATCACTTCGGTGGTAATGCCGGTAATTTGACAATATTTGGTGAAAGCGCGGGCGCGCGTGCGGTTTCATTGCTGACAGCTAGTCCGTTAACCAAAAATTTAATCAGCAAAGCTATAATCCAATCTGGAAACGCATTGTCTTCTAGAGCTTTTCAAAGAGACCCCTTACAAAGTGCTAAAGCTTTAGCCAGGAGTTTGGGCTGTGAAGCAGAAGACGTTGATGAAATTCTAGAGTTTTTAATCGCGACTCCAGCCAAAGACTTGGTTGAGGCAGACGAGAAGTTGAATTCTTTGCAGAAAGTTCTAGAAACGAGTAATAATTTGTTTGGGCTGGTAATCGAGAAGGAGTTTCCAGGAGTCGAAGCTGTCATCAGTGAGCCTTTCATTAACATACTAACATCAGGTAGAACAGCCAACATCCCAATACTAGTTGGAACTACGTCATTAGAGTACGCCTGCGAGAGAAAGACTGATGATCTTCAAGAGTTGATACCAGCTGACCTGAACATAGACAGAAACTCAGAAGAAGCCTTGGCAATCGTAGAAGAAATAAAGAAATTGTATTTCAAAGGGAACCACACAGGAGTGGAGAGTCTACCAGAATACTTCCATCTCCTATCAGACAAGTTGATCAATTTGGACACGCATCGATACATAAAATATTTGCTCCAAGTTACGAACCGTCCAATTTATTACTATAGATTTGATTATGTGGGCGAATTGAATATCGCCCAGAAGATCTTCTTTAGTCTTGGACTGAAGTACGCTATGCACATGGACGAGCTGGGCTATCTGTTCAAGAATGACTTCCAGAAAGACGTGGAGCCGACTCCTGAAGATATCAAGATGCGTGAGAGGATTGTCAGGCTCTGGACTAATTTCGCTAAATCTGGGAACCCGATACCGGACGAAAACCACTATTTGAACACCAACTGGCTACCGGTAACAAACGACAATTTATACTGTCTAAATCTAAATTCAGAATTAACTTTAATCTCAAACCCGGACCAAGAGAAGATGGACTTTTGGGAGAAAATCTACGACAAACACTACAGAATCTGGGATGAAACGACCAACCAGTCTAAAAACGAACCACAAATCGATACATTCAAAAAAGAACCAGAAATCGATACATACCAAAATGAATCAGAAATCGATACATGCAAAAAAGAAACAGAAATCGATACATACAAAAATGAACCAGAAATCGATGCGCCCAAACATGAACCGGAAATAGTGTCCGTTATCGAGACAGTGGTCATAACCCAAGTCATGAACAAAGAGACCAGCGTTTCCGAAGAGAGGACAGAAGTCGTGAGTGACTCTGACCAAATCGACAGAGCCGAAGTTGAAACGCAAATCACAACTACCACACTCCTTGAAAACAACGATGGCTCCCAAACTTTGATAGTCACAGAGAAAACTGAGACCCCCGAACAAGCACCGAGCGTCAAAGAAATGATACTGGAATTCAGTGAACCGGCCCAACTAGAGTCGAATGAACACAAACCGGTCAATGACCACATAAACACTAACGGAGTCGACAAGAAAGCCAGGACTTCGAACGAAATCAAAATGGTGCATAACAGCAACGGTGCTCCAAAGGACGTGATCAGAGCTAATGATCCGCCCGAAGACGATCTACCCAAGAACATAGGAGTGAATAAATTCGTCAGTTTCTTCGAATCGCTAGGCGGAAAGAAGTAA
Protein
MISAVNEFLDDLRGGRMSESPLVTVEQGQLQGRIVNSPSGKAFYSFQGIPYAKPPLGSLRFKAPKPAETWEGIRDATAEGNIAPQIDTSLTKTYTGDENCLYLNVYTPHIAGNLPVMIWIHGGAFKWGSGNETLYGPDYLVEKDVVVVTLNYRCGPLGFLCLNTPEVPGNAGLKDVVQALRWLKQNIKSFGGDPDNFTVFGQSAGGAIVTILTASPLSKNMINKAIVQSGTGISKWAVQNEPLTCAKALASHLGCEADNVDEVLEFLNTVTAKELVEATEIVNSFDSLVVDQNNFFSVVVEKEFPGVEAVLTEPLLDFLTSGRTAEIPIMIGSTTLELLTNLRPSDLQMFIPSDLNIEKDSDESLAIAENLKGLYFTSDTEENKAEGLNRLHSDLLLNININRYIKYLVQNSNQPIYFYKFDYVGQFNFAHKSFFDTELKYALHMDDLGYLFKNDFQKDVDDPSPQDVKMRERMVRLWTNFAKFGNPTPEENHYLPTKWLPVTNDTLYYLNLGQELNLLQNPDEEIMKFWEDLYSKHFKIWEHTKINTIEPPRKLVYPDNEATEEQASIDDGQPSLEEKVVFEEIVPISSVLNPPVNGDAKSNADFKPKISNEIKMAQTSAPKDVVRASDPPEDDLPKNIGAPQPPEPWDGIRDATAEGNVCAQIDPVFAKSYVGDENCLFLNVYTPSTDGAFLPVMIWIHGGGFKWGSGNTNLYGPDFLVDRDVVVVTINYRCGALGFLSLNTPEVPGNAGIKDIVQAIRWVKDNIHHFGGNAGNLTIFGESAGARAVSLLTASPLTKNLISKAIIQSGNALSSRAFQRDPLQSAKALARSLGCEAEDVDEILEFLIATPAKDLVEADEKLNSLQKVLETSNNLFGLVIEKEFPGVEAVISEPFINILTSGRTANIPILVGTTSLEYACERKTDDLQELIPADLNIDRNSEEALAIVEEIKKLYFKGNHTGVESLPEYFHLLSDKLINLDTHRYIKYLLQVTNRPIYYYRFDYVGELNIAQKIFFSLGLKYAMHMDELGYLFKNDFQKDVEPTPEDIKMRERIVRLWTNFAKSGNPIPDENHYLNTNWLPVTNDNLYCLNLNSELTLISNPDQEKMDFWEKIYDKHYRIWDETTNQSKNEPQIDTFKKEPEIDTYQNESEIDTCKKETEIDTYKNEPEIDAPKHEPEIVSVIETVVITQVMNKETSVSEERTEVVSDSDQIDRAEVETQITTTTLLENNDGSQTLIVTEKTETPEQAPSVKEMILEFSEPAQLESNEHKPVNDHINTNGVDKKARTSNEIKMVHNSNGAPKDVIRANDPPEDDLPKNIGVNKFVSFFESLGGKK
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00135 COesterase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
PDB
5IVK
E-value=1.38105e-72,
Score=699
Ontologies
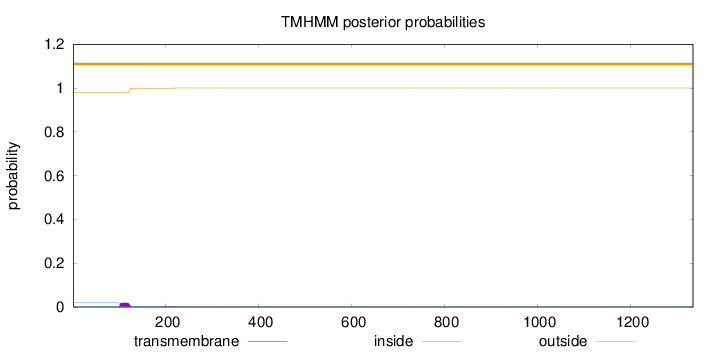
Topology
Length:
1335
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.43273
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.01965
outside
1 - 1335
Population Genetic Test Statistics
Pi
183.282689
Theta
18.939346
Tajima's D
0.385419
CLR
1.136415
CSRT
0.484875756212189
Interpretation
Uncertain
