Gene
KWMTBOMO14071
Pre Gene Modal
BGIBMGA011378
Annotation
PREDICTED:_lysosomal_alpha-mannosidase-like_[Bombyx_mori]
Full name
Alpha-mannosidase
Location in the cell
Nuclear Reliability : 2.12
Sequence
CDS
ATGTGCGTTTTAAACGACCGAGCCCAAGGAGGTACTTCATATAACGACGGAGAAATAGAACTAATGATACATAGAAGACTTCTAACAGATGACGGGTTTGGCTTGGAAGAGAGTCTTAACGAGGAATCTTACGGCACAGGGCTGGTCGTTAGAGGGAAACACAGAATTCTGTTCGGAAACTTCAGACAAGAACTTGAAGAGCTATCGTTCTCGGAGAGAGTATCTCAAGCAGCCAGGCGCTGGAGATATGAACCCTGGTTGTTCTTCGCGCCCGGAGAGAAGATTAGCAGGAGAAAATGGCAGAATGTTAGAAATAAACGTGTAAGTTGA
Protein
MCVLNDRAQGGTSYNDGEIELMIHRRLLTDDGFGLEESLNEESYGTGLVVRGKHRILFGNFRQELEELSFSERVSQAARRWRYEPWLFFAPGEKISRRKWQNVRNKRVS
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the glycosyl hydrolase 38 family.
Feature
chain Alpha-mannosidase
Uniprot
A0A0N0PAB2
A0A194PYQ5
A0A194RQK5
A0A2H1VKW2
A0A2A4IXJ1
A0A3S2NMP6
+ More
Q178W1 A0A182GN04 U5EV98 A0A2J7RN33 A0A2J7RN15 A0A067R124 T1PI90 A0A1J1I5Z3 A0A1J1I212 A0A2P8Z1T1 A0A0C9RCQ8 A0A182K5Y1 A0A182T218 A0A182VJW0 Q5TS83 A0A182KMH3 A0A1S4GYN8 A0A0M9A0P7 A0A182RK95 A0A2A3EH85 A0A182P6H2 A0A182IBN6 A0A182X605 A0A0C9Q6G3 A0A1I8M2X3 A0A1I8M2X1 A0A182MB09 Q178V9 A0A023EUN2 W5JQI4 A0A182QEX5 A0A084VFG4 A0A182IPC8 A0A182YPT8 A0A1W4WS90 A0A182RK94 A0A182WFK9 Q8IPB7 Q9VKV1 A0A1Q3FMS6 A0A1Q3FMY3 A0A087ZZ79 E2ABS8 B0X972 A0A2P8ZEB4 A0A182NTB7 B0X973 A0A154P4N7 A0A023EWA7 B4HWQ4 A0A023EX89 B4Q9B2 A0A0J9R086 A0A182GN05 A0A0R1DR73 A0A3G5BIG9 T1DKM1 A0A2P8Y345 Q178W0 A0A182KHG8 A0A182UGZ7 A0A1S4FBS2 A0A0C9RC73 A0A0L0BRZ6 B3N5F1 A0A0Q5WN53 A0A0C9RFA7 A0A212EWS5 A0A1Q3FJU6 A0A1B6DL32 B0X971 A0A2M4BBR5 A0A2M4A654 A0A2M4BBT1 A0A2M4BB44 A0A2M4BB57 A0A0M4ER29 A0A2M4BAP9 B4MUA9 E2B6B9 A0A232FFB0 K7J2I6 V5HKQ1 A0A1W4UUP0 A0A1J1IUF7 A0A182RK97
Q178W1 A0A182GN04 U5EV98 A0A2J7RN33 A0A2J7RN15 A0A067R124 T1PI90 A0A1J1I5Z3 A0A1J1I212 A0A2P8Z1T1 A0A0C9RCQ8 A0A182K5Y1 A0A182T218 A0A182VJW0 Q5TS83 A0A182KMH3 A0A1S4GYN8 A0A0M9A0P7 A0A182RK95 A0A2A3EH85 A0A182P6H2 A0A182IBN6 A0A182X605 A0A0C9Q6G3 A0A1I8M2X3 A0A1I8M2X1 A0A182MB09 Q178V9 A0A023EUN2 W5JQI4 A0A182QEX5 A0A084VFG4 A0A182IPC8 A0A182YPT8 A0A1W4WS90 A0A182RK94 A0A182WFK9 Q8IPB7 Q9VKV1 A0A1Q3FMS6 A0A1Q3FMY3 A0A087ZZ79 E2ABS8 B0X972 A0A2P8ZEB4 A0A182NTB7 B0X973 A0A154P4N7 A0A023EWA7 B4HWQ4 A0A023EX89 B4Q9B2 A0A0J9R086 A0A182GN05 A0A0R1DR73 A0A3G5BIG9 T1DKM1 A0A2P8Y345 Q178W0 A0A182KHG8 A0A182UGZ7 A0A1S4FBS2 A0A0C9RC73 A0A0L0BRZ6 B3N5F1 A0A0Q5WN53 A0A0C9RFA7 A0A212EWS5 A0A1Q3FJU6 A0A1B6DL32 B0X971 A0A2M4BBR5 A0A2M4A654 A0A2M4BBT1 A0A2M4BB44 A0A2M4BB57 A0A0M4ER29 A0A2M4BAP9 B4MUA9 E2B6B9 A0A232FFB0 K7J2I6 V5HKQ1 A0A1W4UUP0 A0A1J1IUF7 A0A182RK97
EC Number
3.2.1.-
Pubmed
26354079
17510324
26483478
24845553
29403074
12364791
+ More
20966253 25315136 24945155 20920257 23761445 24438588 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17994087 22936249 17550304 30400621 26108605 22118469 28648823 20075255 25765539
20966253 25315136 24945155 20920257 23761445 24438588 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17994087 22936249 17550304 30400621 26108605 22118469 28648823 20075255 25765539
EMBL
KQ459002
KPJ04400.1
KQ459593
KPI96285.1
KQ459875
KPJ19679.1
+ More
ODYU01002940 SOQ41072.1 NWSH01004815 PCG64677.1 RSAL01006410 RVE39916.1 CH477358 EAT42738.1 JXUM01075504 KQ562889 KXJ74904.1 GANO01001972 JAB57899.1 NEVH01002545 PNF42229.1 PNF42230.1 KK853110 KDR11229.1 KA647638 AFP62267.1 CVRI01000038 CRK93801.1 CRK93802.1 PYGN01000238 PSN50451.1 GBYB01014359 JAG84126.1 AAAB01008944 EAL40242.3 KQ435775 KOX74960.1 KZ288262 PBC30371.1 APCN01000712 APCN01000713 APCN01000714 APCN01000715 GBYB01009698 JAG79465.1 AXCM01004170 EAT42740.1 GAPW01000926 JAC12672.1 ADMH02000752 ETN65165.1 AXCN02000716 ATLV01012389 KE524787 KFB36708.1 AE014134 AAN10754.1 AF145601 AAD38576.1 AAF52958.2 GFDL01006180 JAV28865.1 GFDL01006179 JAV28866.1 GL438356 EFN69105.1 DS232521 EDS42978.1 PYGN01000082 PSN54844.1 EDS42979.1 KQ434813 KZC06792.1 GAPW01000098 JAC13500.1 CH480818 EDW52449.1 GAPW01000077 JAC13521.1 CM000361 CM002910 EDX04554.1 KMY89555.1 KMY89556.1 JXUM01075505 KXJ74905.1 CM000157 KRJ97497.1 MK075175 AYV99578.1 GAMD01000881 JAB00710.1 PYGN01000985 PSN38683.1 EAT42739.1 GBYB01004536 JAG74303.1 JRES01001455 KNC22796.1 CH954177 EDV59030.2 KQS70602.1 GBYB01011777 JAG81544.1 AGBW02011898 OWR45950.1 GFDL01007289 JAV27756.1 GEDC01010921 JAS26377.1 EDS42977.1 GGFJ01001332 MBW50473.1 GGFK01002914 MBW36235.1 GGFJ01001331 MBW50472.1 GGFJ01001133 MBW50274.1 GGFJ01001132 MBW50273.1 CP012523 ALC39282.1 GGFJ01000993 MBW50134.1 CH963857 EDW76035.2 GL445930 EFN88812.1 NNAY01000289 OXU29444.1 GANP01008576 JAB75892.1 CVRI01000057 CRL02177.1
ODYU01002940 SOQ41072.1 NWSH01004815 PCG64677.1 RSAL01006410 RVE39916.1 CH477358 EAT42738.1 JXUM01075504 KQ562889 KXJ74904.1 GANO01001972 JAB57899.1 NEVH01002545 PNF42229.1 PNF42230.1 KK853110 KDR11229.1 KA647638 AFP62267.1 CVRI01000038 CRK93801.1 CRK93802.1 PYGN01000238 PSN50451.1 GBYB01014359 JAG84126.1 AAAB01008944 EAL40242.3 KQ435775 KOX74960.1 KZ288262 PBC30371.1 APCN01000712 APCN01000713 APCN01000714 APCN01000715 GBYB01009698 JAG79465.1 AXCM01004170 EAT42740.1 GAPW01000926 JAC12672.1 ADMH02000752 ETN65165.1 AXCN02000716 ATLV01012389 KE524787 KFB36708.1 AE014134 AAN10754.1 AF145601 AAD38576.1 AAF52958.2 GFDL01006180 JAV28865.1 GFDL01006179 JAV28866.1 GL438356 EFN69105.1 DS232521 EDS42978.1 PYGN01000082 PSN54844.1 EDS42979.1 KQ434813 KZC06792.1 GAPW01000098 JAC13500.1 CH480818 EDW52449.1 GAPW01000077 JAC13521.1 CM000361 CM002910 EDX04554.1 KMY89555.1 KMY89556.1 JXUM01075505 KXJ74905.1 CM000157 KRJ97497.1 MK075175 AYV99578.1 GAMD01000881 JAB00710.1 PYGN01000985 PSN38683.1 EAT42739.1 GBYB01004536 JAG74303.1 JRES01001455 KNC22796.1 CH954177 EDV59030.2 KQS70602.1 GBYB01011777 JAG81544.1 AGBW02011898 OWR45950.1 GFDL01007289 JAV27756.1 GEDC01010921 JAS26377.1 EDS42977.1 GGFJ01001332 MBW50473.1 GGFK01002914 MBW36235.1 GGFJ01001331 MBW50472.1 GGFJ01001133 MBW50274.1 GGFJ01001132 MBW50273.1 CP012523 ALC39282.1 GGFJ01000993 MBW50134.1 CH963857 EDW76035.2 GL445930 EFN88812.1 NNAY01000289 OXU29444.1 GANP01008576 JAB75892.1 CVRI01000057 CRL02177.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000283053
UP000008820
UP000069940
+ More
UP000249989 UP000235965 UP000027135 UP000183832 UP000245037 UP000075881 UP000075901 UP000075903 UP000007062 UP000075882 UP000053105 UP000075900 UP000242457 UP000075885 UP000075840 UP000076407 UP000095301 UP000075883 UP000000673 UP000075886 UP000030765 UP000075880 UP000076408 UP000192223 UP000075920 UP000000803 UP000005203 UP000000311 UP000002320 UP000075884 UP000076502 UP000001292 UP000000304 UP000002282 UP000075902 UP000037069 UP000008711 UP000007151 UP000092553 UP000007798 UP000008237 UP000215335 UP000002358 UP000192221
UP000249989 UP000235965 UP000027135 UP000183832 UP000245037 UP000075881 UP000075901 UP000075903 UP000007062 UP000075882 UP000053105 UP000075900 UP000242457 UP000075885 UP000075840 UP000076407 UP000095301 UP000075883 UP000000673 UP000075886 UP000030765 UP000075880 UP000076408 UP000192223 UP000075920 UP000000803 UP000005203 UP000000311 UP000002320 UP000075884 UP000076502 UP000001292 UP000000304 UP000002282 UP000075902 UP000037069 UP000008711 UP000007151 UP000092553 UP000007798 UP000008237 UP000215335 UP000002358 UP000192221
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A0N0PAB2
A0A194PYQ5
A0A194RQK5
A0A2H1VKW2
A0A2A4IXJ1
A0A3S2NMP6
+ More
Q178W1 A0A182GN04 U5EV98 A0A2J7RN33 A0A2J7RN15 A0A067R124 T1PI90 A0A1J1I5Z3 A0A1J1I212 A0A2P8Z1T1 A0A0C9RCQ8 A0A182K5Y1 A0A182T218 A0A182VJW0 Q5TS83 A0A182KMH3 A0A1S4GYN8 A0A0M9A0P7 A0A182RK95 A0A2A3EH85 A0A182P6H2 A0A182IBN6 A0A182X605 A0A0C9Q6G3 A0A1I8M2X3 A0A1I8M2X1 A0A182MB09 Q178V9 A0A023EUN2 W5JQI4 A0A182QEX5 A0A084VFG4 A0A182IPC8 A0A182YPT8 A0A1W4WS90 A0A182RK94 A0A182WFK9 Q8IPB7 Q9VKV1 A0A1Q3FMS6 A0A1Q3FMY3 A0A087ZZ79 E2ABS8 B0X972 A0A2P8ZEB4 A0A182NTB7 B0X973 A0A154P4N7 A0A023EWA7 B4HWQ4 A0A023EX89 B4Q9B2 A0A0J9R086 A0A182GN05 A0A0R1DR73 A0A3G5BIG9 T1DKM1 A0A2P8Y345 Q178W0 A0A182KHG8 A0A182UGZ7 A0A1S4FBS2 A0A0C9RC73 A0A0L0BRZ6 B3N5F1 A0A0Q5WN53 A0A0C9RFA7 A0A212EWS5 A0A1Q3FJU6 A0A1B6DL32 B0X971 A0A2M4BBR5 A0A2M4A654 A0A2M4BBT1 A0A2M4BB44 A0A2M4BB57 A0A0M4ER29 A0A2M4BAP9 B4MUA9 E2B6B9 A0A232FFB0 K7J2I6 V5HKQ1 A0A1W4UUP0 A0A1J1IUF7 A0A182RK97
Q178W1 A0A182GN04 U5EV98 A0A2J7RN33 A0A2J7RN15 A0A067R124 T1PI90 A0A1J1I5Z3 A0A1J1I212 A0A2P8Z1T1 A0A0C9RCQ8 A0A182K5Y1 A0A182T218 A0A182VJW0 Q5TS83 A0A182KMH3 A0A1S4GYN8 A0A0M9A0P7 A0A182RK95 A0A2A3EH85 A0A182P6H2 A0A182IBN6 A0A182X605 A0A0C9Q6G3 A0A1I8M2X3 A0A1I8M2X1 A0A182MB09 Q178V9 A0A023EUN2 W5JQI4 A0A182QEX5 A0A084VFG4 A0A182IPC8 A0A182YPT8 A0A1W4WS90 A0A182RK94 A0A182WFK9 Q8IPB7 Q9VKV1 A0A1Q3FMS6 A0A1Q3FMY3 A0A087ZZ79 E2ABS8 B0X972 A0A2P8ZEB4 A0A182NTB7 B0X973 A0A154P4N7 A0A023EWA7 B4HWQ4 A0A023EX89 B4Q9B2 A0A0J9R086 A0A182GN05 A0A0R1DR73 A0A3G5BIG9 T1DKM1 A0A2P8Y345 Q178W0 A0A182KHG8 A0A182UGZ7 A0A1S4FBS2 A0A0C9RC73 A0A0L0BRZ6 B3N5F1 A0A0Q5WN53 A0A0C9RFA7 A0A212EWS5 A0A1Q3FJU6 A0A1B6DL32 B0X971 A0A2M4BBR5 A0A2M4A654 A0A2M4BBT1 A0A2M4BB44 A0A2M4BB57 A0A0M4ER29 A0A2M4BAP9 B4MUA9 E2B6B9 A0A232FFB0 K7J2I6 V5HKQ1 A0A1W4UUP0 A0A1J1IUF7 A0A182RK97
PDB
1O7D
E-value=2.80715e-11,
Score=158
Ontologies
GO
Topology
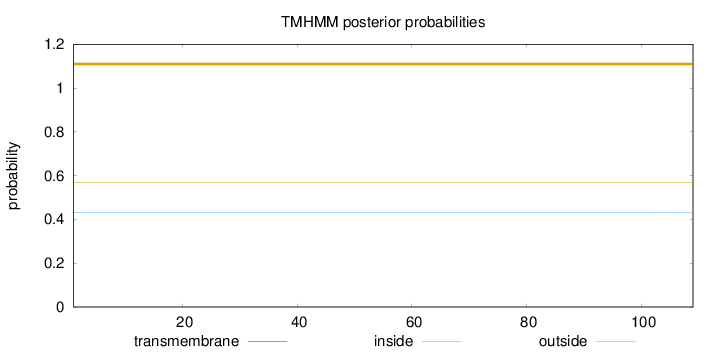
Length:
109
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.43074
outside
1 - 109
Population Genetic Test Statistics
Pi
338.804219
Theta
175.594449
Tajima's D
2.92593
CLR
0.383802
CSRT
0.975201239938003
Interpretation
Uncertain
