Gene
KWMTBOMO14068
Annotation
PREDICTED:_lysosomal_alpha-mannosidase-like_[Papilio_xuthus]
Full name
Alpha-mannosidase
Location in the cell
Mitochondrial Reliability : 1.662
Sequence
CDS
ATGCGCTTCTTCCATATTGGAAAGCATTCATTCACCTTCTCCGAAACGTCGTATTTCTGGCGCTGGTGGAAAGAACAGCGTCCTGTAACGAGGTCCACCTTCCGTAGCCTCGTGCAGGAAGGTCGCGTGGAGTTCGCCGGAGGAGGCTGGGTCCAGAACGACGAAGCGACTTCCGATTATCTCCAGATCATTGACCAATATACTTGGGGATTGAGGAAACTGAACGACACTTTAGGCCTGTGTGGTAAGCCTAAAGCAGCTTGGCAGATTGACACATATGGCCACACCAGGGAACAGGCATCTTTACTGGCACAAATGGGCTTTGATGGCTTATTTATCGGTACATATCTCGGTTTTTATTTACTTATCGCTTAA
Protein
MRFFHIGKHSFTFSETSYFWRWWKEQRPVTRSTFRSLVQEGRVEFAGGGWVQNDEATSDYLQIIDQYTWGLRKLNDTLGLCGKPKAAWQIDTYGHTREQASLLAQMGFDGLFIGTYLGFYLLIA
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the glycosyl hydrolase 38 family.
Feature
chain Alpha-mannosidase
Uniprot
A0A2A4IWX3
A0A2A4IXJ1
A0A194PYQ5
A0A194RQK5
A0A2W1BMA0
A0A2H1VKW2
+ More
E0VRZ4 A0A1Y1KHF4 Q178V9 A0A182GN05 U5EV98 A0A023EX89 A0A3G5BIG9 A0A0A1WJW5 A0A2P8Y360 A0A1L8DR39 A0A2A4JTC5 A0A146LUI9 A0A0A9VXB9 A0A034V372 A0A0A1WZV2 A0A1L8DQQ1 W8APD9 A0A0A1XEF2 A0A2A4JU30 A0A0P5C6A1 A0A2P8Y345 A0A0P5CFQ9 A0A1S4FBS2 D6WI18 A0A1B0CB94 A0A2A3EH85 B3MJI9 A0A3G5BIM3 A0A0P8XGR3 U4V0C3 A0A2P8Z1T1 A0A0P5C669 J9JL57 A0A2H8TPV8 A0A1J1I5Z3 A0A1J1I212 A0A0P5C6H0 Q178W1 A0A1Y1KKT1 A0A3B0JHP2 A0A3B0K7V8 A0A3B0JH41 N6TNX4 A0A0Q9XIT5 B4KHR0 B3N5F1 A0A087T0N6 A0A0Q5WN53 E9H264 A0A2H1VPQ4 U4TT01 A0A154P4N7 U4TTZ9 N6ULX5 A0A023EWA7 B4MUA9 A0A1B6CNQ5 A0A212F5R4 A0A0L7QJS0 A0A3Q0J7L7 A0A2M4BAP9 A0A182QEX5 A0A1B0GPJ0 A0A0R1DR73 A0A1B6CQR4 Q8IPB7 B4Q9B2 Q9VKV1 B4HWQ4 A0A0J9R086 A0A2I4CBX4 A0A2J7RN15 A0A2P8ZEB4 A0A182P6H2 A0A0M4ER29 A0A2J7RN33 A0A336LVA7 A0A1I8QD07 A0A336LYZ7 A0A1W3JCU7 A0A087ZZ79 A0A0K8WDF9 A0A182RK94 A0A1I8QCY1 Q178W0 A0A2L2Y690 A0A0K8UEK7 A0A1J1IUF7 A0A1A9W6T1 A0A0K8U3N4 A0A0K8VT60 A0A146VDZ7 A0A182NTB7 A0A0K8VTI9
E0VRZ4 A0A1Y1KHF4 Q178V9 A0A182GN05 U5EV98 A0A023EX89 A0A3G5BIG9 A0A0A1WJW5 A0A2P8Y360 A0A1L8DR39 A0A2A4JTC5 A0A146LUI9 A0A0A9VXB9 A0A034V372 A0A0A1WZV2 A0A1L8DQQ1 W8APD9 A0A0A1XEF2 A0A2A4JU30 A0A0P5C6A1 A0A2P8Y345 A0A0P5CFQ9 A0A1S4FBS2 D6WI18 A0A1B0CB94 A0A2A3EH85 B3MJI9 A0A3G5BIM3 A0A0P8XGR3 U4V0C3 A0A2P8Z1T1 A0A0P5C669 J9JL57 A0A2H8TPV8 A0A1J1I5Z3 A0A1J1I212 A0A0P5C6H0 Q178W1 A0A1Y1KKT1 A0A3B0JHP2 A0A3B0K7V8 A0A3B0JH41 N6TNX4 A0A0Q9XIT5 B4KHR0 B3N5F1 A0A087T0N6 A0A0Q5WN53 E9H264 A0A2H1VPQ4 U4TT01 A0A154P4N7 U4TTZ9 N6ULX5 A0A023EWA7 B4MUA9 A0A1B6CNQ5 A0A212F5R4 A0A0L7QJS0 A0A3Q0J7L7 A0A2M4BAP9 A0A182QEX5 A0A1B0GPJ0 A0A0R1DR73 A0A1B6CQR4 Q8IPB7 B4Q9B2 Q9VKV1 B4HWQ4 A0A0J9R086 A0A2I4CBX4 A0A2J7RN15 A0A2P8ZEB4 A0A182P6H2 A0A0M4ER29 A0A2J7RN33 A0A336LVA7 A0A1I8QD07 A0A336LYZ7 A0A1W3JCU7 A0A087ZZ79 A0A0K8WDF9 A0A182RK94 A0A1I8QCY1 Q178W0 A0A2L2Y690 A0A0K8UEK7 A0A1J1IUF7 A0A1A9W6T1 A0A0K8U3N4 A0A0K8VT60 A0A146VDZ7 A0A182NTB7 A0A0K8VTI9
EC Number
3.2.1.-
Pubmed
26354079
28756777
20566863
28004739
17510324
26483478
+ More
24945155 30400621 25830018 29403074 26823975 25401762 25348373 24495485 18362917 19820115 17994087 23537049 21292972 22118469 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 26109357 26109356 26561354
24945155 30400621 25830018 29403074 26823975 25401762 25348373 24495485 18362917 19820115 17994087 23537049 21292972 22118469 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 26109357 26109356 26561354
EMBL
NWSH01006168
PCG63620.1
NWSH01004815
PCG64677.1
KQ459593
KPI96285.1
+ More
KQ459875 KPJ19679.1 KZ150153 PZC72813.1 ODYU01002940 SOQ41072.1 DS235639 EEB16150.1 GEZM01085309 JAV60008.1 CH477358 EAT42740.1 JXUM01075505 KQ562889 KXJ74905.1 GANO01001972 JAB57899.1 GAPW01000077 JAC13521.1 MK075175 AYV99578.1 GBXI01014983 JAC99308.1 PYGN01000985 PSN38671.1 GFDF01005289 JAV08795.1 NWSH01000615 PCG75287.1 GDHC01008237 JAQ10392.1 GBHO01043763 GBHO01043759 JAF99840.1 JAF99844.1 GAKP01021246 JAC37706.1 GBXI01009935 JAD04357.1 GFDF01005288 JAV08796.1 GAMC01015917 GAMC01015916 GAMC01015915 GAMC01015914 JAB90640.1 GBXI01004996 JAD09296.1 PCG75286.1 GDIP01174920 JAJ48482.1 PSN38683.1 GDIP01174916 JAJ48486.1 KQ971332 EEZ99703.1 AJWK01004914 KZ288262 PBC30371.1 CH902620 EDV32357.1 MK075237 AYV99640.1 KPU73915.1 KI209786 ERL96095.1 PYGN01000238 PSN50451.1 GDIP01174918 JAJ48484.1 ABLF02026049 GFXV01004418 MBW16223.1 CVRI01000038 CRK93801.1 CRK93802.1 GDIP01174919 JAJ48483.1 EAT42738.1 GEZM01085310 JAV60006.1 OUUW01000006 SPP81715.1 SPP81716.1 SPP81714.1 APGK01026882 KB740648 ENN79713.1 CH933807 KRG03704.1 EDW13347.1 CH954177 EDV59030.2 KK112841 KFM58675.1 KQS70602.1 GL732585 EFX74109.1 ODYU01003694 SOQ42778.1 KB630779 ERL83857.1 KQ434813 KZC06792.1 KB631357 ERL84262.1 ENN79712.1 GAPW01000098 JAC13500.1 CH963857 EDW76035.2 GEDC01022285 JAS15013.1 AGBW02010159 OWR49049.1 KQ415041 KOC58867.1 GGFJ01000993 MBW50134.1 AXCN02000716 AJVK01005894 AJVK01005895 CM000157 KRJ97497.1 GEDC01021511 JAS15787.1 AE014134 AAN10754.1 CM000361 CM002910 EDX04554.1 KMY89555.1 AF145601 AAD38576.1 AAF52958.2 CH480818 EDW52449.1 KMY89556.1 NEVH01002545 PNF42230.1 PYGN01000082 PSN54844.1 CP012523 ALC39282.1 PNF42229.1 UFQS01000218 UFQT01000218 SSX01504.1 SSX21884.1 SSX01503.1 SSX21883.1 GDHF01003182 JAI49132.1 EAT42739.1 IAAA01017254 LAA03679.1 GDHF01027225 GDHF01012263 JAI25089.1 JAI40051.1 CVRI01000057 CRL02177.1 GDHF01030990 JAI21324.1 GDHF01010271 GDHF01002021 JAI42043.1 JAI50293.1 GCES01071146 JAR15177.1 GDHF01010454 JAI41860.1
KQ459875 KPJ19679.1 KZ150153 PZC72813.1 ODYU01002940 SOQ41072.1 DS235639 EEB16150.1 GEZM01085309 JAV60008.1 CH477358 EAT42740.1 JXUM01075505 KQ562889 KXJ74905.1 GANO01001972 JAB57899.1 GAPW01000077 JAC13521.1 MK075175 AYV99578.1 GBXI01014983 JAC99308.1 PYGN01000985 PSN38671.1 GFDF01005289 JAV08795.1 NWSH01000615 PCG75287.1 GDHC01008237 JAQ10392.1 GBHO01043763 GBHO01043759 JAF99840.1 JAF99844.1 GAKP01021246 JAC37706.1 GBXI01009935 JAD04357.1 GFDF01005288 JAV08796.1 GAMC01015917 GAMC01015916 GAMC01015915 GAMC01015914 JAB90640.1 GBXI01004996 JAD09296.1 PCG75286.1 GDIP01174920 JAJ48482.1 PSN38683.1 GDIP01174916 JAJ48486.1 KQ971332 EEZ99703.1 AJWK01004914 KZ288262 PBC30371.1 CH902620 EDV32357.1 MK075237 AYV99640.1 KPU73915.1 KI209786 ERL96095.1 PYGN01000238 PSN50451.1 GDIP01174918 JAJ48484.1 ABLF02026049 GFXV01004418 MBW16223.1 CVRI01000038 CRK93801.1 CRK93802.1 GDIP01174919 JAJ48483.1 EAT42738.1 GEZM01085310 JAV60006.1 OUUW01000006 SPP81715.1 SPP81716.1 SPP81714.1 APGK01026882 KB740648 ENN79713.1 CH933807 KRG03704.1 EDW13347.1 CH954177 EDV59030.2 KK112841 KFM58675.1 KQS70602.1 GL732585 EFX74109.1 ODYU01003694 SOQ42778.1 KB630779 ERL83857.1 KQ434813 KZC06792.1 KB631357 ERL84262.1 ENN79712.1 GAPW01000098 JAC13500.1 CH963857 EDW76035.2 GEDC01022285 JAS15013.1 AGBW02010159 OWR49049.1 KQ415041 KOC58867.1 GGFJ01000993 MBW50134.1 AXCN02000716 AJVK01005894 AJVK01005895 CM000157 KRJ97497.1 GEDC01021511 JAS15787.1 AE014134 AAN10754.1 CM000361 CM002910 EDX04554.1 KMY89555.1 AF145601 AAD38576.1 AAF52958.2 CH480818 EDW52449.1 KMY89556.1 NEVH01002545 PNF42230.1 PYGN01000082 PSN54844.1 CP012523 ALC39282.1 PNF42229.1 UFQS01000218 UFQT01000218 SSX01504.1 SSX21884.1 SSX01503.1 SSX21883.1 GDHF01003182 JAI49132.1 EAT42739.1 IAAA01017254 LAA03679.1 GDHF01027225 GDHF01012263 JAI25089.1 JAI40051.1 CVRI01000057 CRL02177.1 GDHF01030990 JAI21324.1 GDHF01010271 GDHF01002021 JAI42043.1 JAI50293.1 GCES01071146 JAR15177.1 GDHF01010454 JAI41860.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000009046
UP000008820
UP000069940
+ More
UP000249989 UP000245037 UP000007266 UP000092461 UP000242457 UP000007801 UP000030742 UP000007819 UP000183832 UP000268350 UP000019118 UP000009192 UP000008711 UP000054359 UP000000305 UP000076502 UP000007798 UP000007151 UP000053825 UP000079169 UP000075886 UP000092462 UP000002282 UP000000803 UP000000304 UP000001292 UP000192220 UP000235965 UP000075885 UP000092553 UP000095300 UP000005203 UP000075900 UP000091820 UP000075884
UP000249989 UP000245037 UP000007266 UP000092461 UP000242457 UP000007801 UP000030742 UP000007819 UP000183832 UP000268350 UP000019118 UP000009192 UP000008711 UP000054359 UP000000305 UP000076502 UP000007798 UP000007151 UP000053825 UP000079169 UP000075886 UP000092462 UP000002282 UP000000803 UP000000304 UP000001292 UP000192220 UP000235965 UP000075885 UP000092553 UP000095300 UP000005203 UP000075900 UP000091820 UP000075884
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2A4IWX3
A0A2A4IXJ1
A0A194PYQ5
A0A194RQK5
A0A2W1BMA0
A0A2H1VKW2
+ More
E0VRZ4 A0A1Y1KHF4 Q178V9 A0A182GN05 U5EV98 A0A023EX89 A0A3G5BIG9 A0A0A1WJW5 A0A2P8Y360 A0A1L8DR39 A0A2A4JTC5 A0A146LUI9 A0A0A9VXB9 A0A034V372 A0A0A1WZV2 A0A1L8DQQ1 W8APD9 A0A0A1XEF2 A0A2A4JU30 A0A0P5C6A1 A0A2P8Y345 A0A0P5CFQ9 A0A1S4FBS2 D6WI18 A0A1B0CB94 A0A2A3EH85 B3MJI9 A0A3G5BIM3 A0A0P8XGR3 U4V0C3 A0A2P8Z1T1 A0A0P5C669 J9JL57 A0A2H8TPV8 A0A1J1I5Z3 A0A1J1I212 A0A0P5C6H0 Q178W1 A0A1Y1KKT1 A0A3B0JHP2 A0A3B0K7V8 A0A3B0JH41 N6TNX4 A0A0Q9XIT5 B4KHR0 B3N5F1 A0A087T0N6 A0A0Q5WN53 E9H264 A0A2H1VPQ4 U4TT01 A0A154P4N7 U4TTZ9 N6ULX5 A0A023EWA7 B4MUA9 A0A1B6CNQ5 A0A212F5R4 A0A0L7QJS0 A0A3Q0J7L7 A0A2M4BAP9 A0A182QEX5 A0A1B0GPJ0 A0A0R1DR73 A0A1B6CQR4 Q8IPB7 B4Q9B2 Q9VKV1 B4HWQ4 A0A0J9R086 A0A2I4CBX4 A0A2J7RN15 A0A2P8ZEB4 A0A182P6H2 A0A0M4ER29 A0A2J7RN33 A0A336LVA7 A0A1I8QD07 A0A336LYZ7 A0A1W3JCU7 A0A087ZZ79 A0A0K8WDF9 A0A182RK94 A0A1I8QCY1 Q178W0 A0A2L2Y690 A0A0K8UEK7 A0A1J1IUF7 A0A1A9W6T1 A0A0K8U3N4 A0A0K8VT60 A0A146VDZ7 A0A182NTB7 A0A0K8VTI9
E0VRZ4 A0A1Y1KHF4 Q178V9 A0A182GN05 U5EV98 A0A023EX89 A0A3G5BIG9 A0A0A1WJW5 A0A2P8Y360 A0A1L8DR39 A0A2A4JTC5 A0A146LUI9 A0A0A9VXB9 A0A034V372 A0A0A1WZV2 A0A1L8DQQ1 W8APD9 A0A0A1XEF2 A0A2A4JU30 A0A0P5C6A1 A0A2P8Y345 A0A0P5CFQ9 A0A1S4FBS2 D6WI18 A0A1B0CB94 A0A2A3EH85 B3MJI9 A0A3G5BIM3 A0A0P8XGR3 U4V0C3 A0A2P8Z1T1 A0A0P5C669 J9JL57 A0A2H8TPV8 A0A1J1I5Z3 A0A1J1I212 A0A0P5C6H0 Q178W1 A0A1Y1KKT1 A0A3B0JHP2 A0A3B0K7V8 A0A3B0JH41 N6TNX4 A0A0Q9XIT5 B4KHR0 B3N5F1 A0A087T0N6 A0A0Q5WN53 E9H264 A0A2H1VPQ4 U4TT01 A0A154P4N7 U4TTZ9 N6ULX5 A0A023EWA7 B4MUA9 A0A1B6CNQ5 A0A212F5R4 A0A0L7QJS0 A0A3Q0J7L7 A0A2M4BAP9 A0A182QEX5 A0A1B0GPJ0 A0A0R1DR73 A0A1B6CQR4 Q8IPB7 B4Q9B2 Q9VKV1 B4HWQ4 A0A0J9R086 A0A2I4CBX4 A0A2J7RN15 A0A2P8ZEB4 A0A182P6H2 A0A0M4ER29 A0A2J7RN33 A0A336LVA7 A0A1I8QD07 A0A336LYZ7 A0A1W3JCU7 A0A087ZZ79 A0A0K8WDF9 A0A182RK94 A0A1I8QCY1 Q178W0 A0A2L2Y690 A0A0K8UEK7 A0A1J1IUF7 A0A1A9W6T1 A0A0K8U3N4 A0A0K8VT60 A0A146VDZ7 A0A182NTB7 A0A0K8VTI9
PDB
1O7D
E-value=1.24708e-31,
Score=334
Ontologies
KEGG
GO
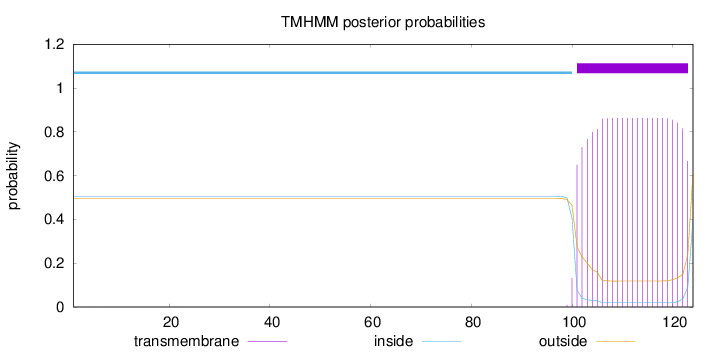
Topology
Length:
124
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.15354
Exp number, first 60 AAs:
0.00297
Total prob of N-in:
0.50509
inside
1 - 100
TMhelix
101 - 123
outside
124 - 124
Population Genetic Test Statistics
Pi
88.456005
Theta
63.915298
Tajima's D
1.198562
CLR
5.578457
CSRT
0.714214289285536
Interpretation
Uncertain
