Gene
KWMTBOMO14066
Pre Gene Modal
BGIBMGA011226
Annotation
PREDICTED:_solute_carrier_family_22_member_13-like_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.85
Sequence
CDS
ATGGGATCACGTGAGAGGAGCTTTCAGGGCACGGAGCCGCTGCCCCATGACCCTACCAACGACTTGGAAATGGTCGAGCGAAAAGAAAACGGTGCTAATCGCCACAGATTGAAGGATCTTGATGACCTTCTGCCGTACATGGGAGAGTTTGGTTGGTACCAGCGCATGCTGTTCTTGTTGATGATACCTTACGCGTTCTTCTTCGCGTTCGTCTACTTCGGGCAGATATTCATGGCCATAACTCCGGAGGAGCACTGGTGCTCTGTCCCGGAATTGGCGAACTTGAGCGTGATTGAACGGCGTCATCTATCCATACCGATGTTAAACAATAAATATTCAAAATGCGAAGTTTACGATGTAGATTGGTCGAAACTGTTACAAGATGGCGTCCGTCAGCCGAACTCAAAATGGCCGACCAAGAAATGTCAAAATAAATGGGAGTTTAATTACACTGATGTTCCTTATGAAACAATTGCAACACAGCAAGGCTGGGTGTGTGACAAGGACAACTATGCCGCCACTGCTCAGTCGGTGTTTTTCTGTGGGGCCATACTGGGTGGATTTATCTTTGGTTGGATAGCTGACAAATATGGTCGTATACCAGCTCTAGTGGGTACGAATGTAGCCGGATTTATTGGCGGAATCGGAACTGTGTTTTCAAATGATTTTTTATCGTTCTGCCTCTGCCGATTCATTGTGGGCTTAGCCTATGACAATTGTTTTATTATCATGTATATCGTCGTGCTGGAGTACGTTGGCCTCAAATGGAGAACCTTCGTGGCGAATATGTCCATAGGAGTATACTTCACGTTTGCAGCGTGCCTCCTACCCTGGATCGCTCTAGCGGTCGCGGACTGGAAGATTTATACTCTGATCATTAGCGTGCCTTTAGCTTTGGCCGTCTTCACTGGCTGCGTCGTACCAGAGAGCGCCAGGTGGTTGATATCGCAAGGGCGGATCGATGAAGCACTAAAAATAATGATGAGGTGCGCGAAGGTCAACGGTAATGCACTACCAGAGGATATTATCAAGGAATTTCACGAAACAGCAACAGGAATAGCTAAAGCCGAACAAGAATGTAAGAACTACAGCGTATTAGATCTATTCAAGACTCCTCGATTGCGAAAGCACAGCATTCTTCTGGTGTTCATTTGGATTCTAATAGCCATGGCTTTCGATGGACACGTCAGGAATGTGGGCACTCTGGGCCTGGACATGTTCCTGACCTTCACTGTGGCTACGGCGACCGAGTTTCCAGCCGATGTTTTGCTTACCCTGGTCTTGGATGTGTGA
Protein
MGSRERSFQGTEPLPHDPTNDLEMVERKENGANRHRLKDLDDLLPYMGEFGWYQRMLFLLMIPYAFFFAFVYFGQIFMAITPEEHWCSVPELANLSVIERRHLSIPMLNNKYSKCEVYDVDWSKLLQDGVRQPNSKWPTKKCQNKWEFNYTDVPYETIATQQGWVCDKDNYAATAQSVFFCGAILGGFIFGWIADKYGRIPALVGTNVAGFIGGIGTVFSNDFLSFCLCRFIVGLAYDNCFIIMYIVVLEYVGLKWRTFVANMSIGVYFTFAACLLPWIALAVADWKIYTLIISVPLALAVFTGCVVPESARWLISQGRIDEALKIMMRCAKVNGNALPEDIIKEFHETATGIAKAEQECKNYSVLDLFKTPRLRKHSILLVFIWILIAMAFDGHVRNVGTLGLDMFLTFTVATATEFPADVLLTLVLDV
Summary
Uniprot
H9JNX2
A0A2W1BJ02
A0A194RU78
A0A194PTI5
A0A212FCX8
A0A3S2PHH3
+ More
A0A2W1BCV5 A0A2H1VWQ6 A0A212FD00 F4W561 E2BDY3 A0A195D0Q3 A0A151J3V4 A0A158NZC1 A0A026W645 A0A154PDD7 A0A194PS69 A0A151WQU6 E2AZ43 A0A194RQU6 E9J4S9 A0A0L7RG38 A0A2A4IX18 D6WHS6 A0A1B6KUA4 A0A1B6FLN6 A0A195FUP4 A0A1Y1LPP5 A0A3B0KFJ9 A0A1I8NGG5 B4MLM1 A0A310SF21 B4L9P9 A0A0A1XP62 A0A0R3P8K9 A0A1J1I7T3 A0A0K8USS0 A0A023F004 A0A0V0GBK0 W8AVR2 A0A0L0BT78 A0A1W4UXC6 B3NCD2 A0A232F2T3 Q9VRT4 A0A151I4F7 A0A0J9RR75 B4PJC0 A0A224XLE8 T1DJX3 A0A0P8YIG7 T1I0F0 A0A0L0C4M2 A0A1Q3G090 A0A3F2YXP2 E0VI57 A0A0P6IYS1 A0A182Y2J8 A0A182LKD6 Q7Q570 A0A182VNM0 A0A182QEZ0 A0A182TJ22 B4LDD7 A0A0M4EFT7 A0A1I8Q048 A0A0K8VFG7 A0A2J7R373 A0A1I8NXE0 A0A1A9Y9S3 A0A2J7R361 A0A1A9VMM4 A0A0J7NSN3 A0A067RU41 A0A1B0GG54 A0A1A9WFE2 T1PHJ0 A0A2A3E4H2 A0A1B0AIC1 A0A0C9QKP2 A0A336K3J5 A0A336JZ48 A0A2A4K3T5 A0A2W1BGI1 A0A1S3D083 A0A1A9ZKZ7 H9JNX1 A0A088A3S5 A0A1A9X3B2 A0A1L8DE47 A0A195D0Q4 A0A0L7RGH8 A0A195FT31 N6TV42 A0A1A9V9X7 A0A088AMR8 A0A1B0BC30 A0A151WQK2 A0A1B0F9D3 A0A310SBQ9
A0A2W1BCV5 A0A2H1VWQ6 A0A212FD00 F4W561 E2BDY3 A0A195D0Q3 A0A151J3V4 A0A158NZC1 A0A026W645 A0A154PDD7 A0A194PS69 A0A151WQU6 E2AZ43 A0A194RQU6 E9J4S9 A0A0L7RG38 A0A2A4IX18 D6WHS6 A0A1B6KUA4 A0A1B6FLN6 A0A195FUP4 A0A1Y1LPP5 A0A3B0KFJ9 A0A1I8NGG5 B4MLM1 A0A310SF21 B4L9P9 A0A0A1XP62 A0A0R3P8K9 A0A1J1I7T3 A0A0K8USS0 A0A023F004 A0A0V0GBK0 W8AVR2 A0A0L0BT78 A0A1W4UXC6 B3NCD2 A0A232F2T3 Q9VRT4 A0A151I4F7 A0A0J9RR75 B4PJC0 A0A224XLE8 T1DJX3 A0A0P8YIG7 T1I0F0 A0A0L0C4M2 A0A1Q3G090 A0A3F2YXP2 E0VI57 A0A0P6IYS1 A0A182Y2J8 A0A182LKD6 Q7Q570 A0A182VNM0 A0A182QEZ0 A0A182TJ22 B4LDD7 A0A0M4EFT7 A0A1I8Q048 A0A0K8VFG7 A0A2J7R373 A0A1I8NXE0 A0A1A9Y9S3 A0A2J7R361 A0A1A9VMM4 A0A0J7NSN3 A0A067RU41 A0A1B0GG54 A0A1A9WFE2 T1PHJ0 A0A2A3E4H2 A0A1B0AIC1 A0A0C9QKP2 A0A336K3J5 A0A336JZ48 A0A2A4K3T5 A0A2W1BGI1 A0A1S3D083 A0A1A9ZKZ7 H9JNX1 A0A088A3S5 A0A1A9X3B2 A0A1L8DE47 A0A195D0Q4 A0A0L7RGH8 A0A195FT31 N6TV42 A0A1A9V9X7 A0A088AMR8 A0A1B0BC30 A0A151WQK2 A0A1B0F9D3 A0A310SBQ9
Pubmed
19121390
28756777
26354079
22118469
21719571
20798317
+ More
21347285 24508170 30249741 21282665 18362917 19820115 28004739 25315136 17994087 25830018 15632085 25474469 24495485 26108605 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 20566863 26999592 25244985 20966253 12364791 14747013 17210077 24845553 23537049
21347285 24508170 30249741 21282665 18362917 19820115 28004739 25315136 17994087 25830018 15632085 25474469 24495485 26108605 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 20566863 26999592 25244985 20966253 12364791 14747013 17210077 24845553 23537049
EMBL
BABH01009800
BABH01009801
KZ150153
PZC72816.1
KQ459875
KPJ19681.1
+ More
KQ459593 KPI96288.1 AGBW02009123 OWR51601.1 RSAL01000031 RVE51608.1 KZ150454 PZC70816.1 ODYU01004909 SOQ45269.1 OWR51600.1 GL887605 EGI70657.1 GL447693 EFN86109.1 KQ977004 KYN06495.1 KQ980249 KYN17104.1 ADTU01004402 ADTU01004403 ADTU01004404 ADTU01004405 KK107419 QOIP01000003 EZA51081.1 RLU24986.1 KQ434878 KZC09853.1 KPI96286.1 KQ982821 KYQ50181.1 GL444080 EFN61301.1 KPJ19680.1 GL768119 EFZ12162.1 KQ414596 KOC69917.1 NWSH01006028 PCG63732.1 KQ971321 EFA00687.1 GEBQ01024996 JAT14981.1 GECZ01018651 JAS51118.1 KQ981276 KYN43599.1 GEZM01052731 JAV74290.1 OUUW01000009 SPP85099.1 CH963847 EDW72947.2 KQ769818 OAD52819.1 CH933816 EDW17097.1 GBXI01001560 JAD12732.1 CH379069 KRT08150.1 CVRI01000042 CRK95636.1 GDHF01022741 JAI29573.1 GBBI01004623 JAC14089.1 GECL01001415 JAP04709.1 GAMC01016163 JAB90392.1 JRES01001371 KNC23290.1 CH954178 EDV51090.2 NNAY01001101 OXU25136.1 AE014296 AAF50706.2 KQ976448 KYM85871.1 CM002912 KMY97849.1 CM000159 EDW93588.2 GFTR01007453 JAW08973.1 GAMD01001231 JAB00360.1 CH902618 KPU78771.1 ACPB03009635 ACPB03009636 ACPB03009637 ACPB03009638 ACPB03009639 JRES01000930 KNC27191.1 GFDL01001826 JAV33219.1 DS235184 EEB13063.1 GDUN01000921 JAN94998.1 AAAB01008960 EAA11678.4 AXCN02000878 CH940647 EDW68875.2 CP012525 ALC43177.1 GDHF01014620 JAI37694.1 NEVH01007822 PNF35273.1 PNF35272.1 LBMM01001988 KMQ95440.1 KK852417 KDR24330.1 CCAG010010771 KA648236 AFP62865.1 KZ288416 PBC26059.1 GBYB01001152 JAG70919.1 UFQS01000037 UFQT01000037 SSW98015.1 SSX18401.1 SSW98016.1 SSX18402.1 NWSH01000178 PCG78709.1 PZC72815.1 BABH01009804 BABH01009805 GFDF01009490 JAV04594.1 KYN06498.1 KOC69919.1 KYN43601.1 APGK01053700 APGK01053701 KB741231 ENN72266.1 JXJN01011785 KYQ50179.1 CCAG010012933 KQ761655 OAD57197.1
KQ459593 KPI96288.1 AGBW02009123 OWR51601.1 RSAL01000031 RVE51608.1 KZ150454 PZC70816.1 ODYU01004909 SOQ45269.1 OWR51600.1 GL887605 EGI70657.1 GL447693 EFN86109.1 KQ977004 KYN06495.1 KQ980249 KYN17104.1 ADTU01004402 ADTU01004403 ADTU01004404 ADTU01004405 KK107419 QOIP01000003 EZA51081.1 RLU24986.1 KQ434878 KZC09853.1 KPI96286.1 KQ982821 KYQ50181.1 GL444080 EFN61301.1 KPJ19680.1 GL768119 EFZ12162.1 KQ414596 KOC69917.1 NWSH01006028 PCG63732.1 KQ971321 EFA00687.1 GEBQ01024996 JAT14981.1 GECZ01018651 JAS51118.1 KQ981276 KYN43599.1 GEZM01052731 JAV74290.1 OUUW01000009 SPP85099.1 CH963847 EDW72947.2 KQ769818 OAD52819.1 CH933816 EDW17097.1 GBXI01001560 JAD12732.1 CH379069 KRT08150.1 CVRI01000042 CRK95636.1 GDHF01022741 JAI29573.1 GBBI01004623 JAC14089.1 GECL01001415 JAP04709.1 GAMC01016163 JAB90392.1 JRES01001371 KNC23290.1 CH954178 EDV51090.2 NNAY01001101 OXU25136.1 AE014296 AAF50706.2 KQ976448 KYM85871.1 CM002912 KMY97849.1 CM000159 EDW93588.2 GFTR01007453 JAW08973.1 GAMD01001231 JAB00360.1 CH902618 KPU78771.1 ACPB03009635 ACPB03009636 ACPB03009637 ACPB03009638 ACPB03009639 JRES01000930 KNC27191.1 GFDL01001826 JAV33219.1 DS235184 EEB13063.1 GDUN01000921 JAN94998.1 AAAB01008960 EAA11678.4 AXCN02000878 CH940647 EDW68875.2 CP012525 ALC43177.1 GDHF01014620 JAI37694.1 NEVH01007822 PNF35273.1 PNF35272.1 LBMM01001988 KMQ95440.1 KK852417 KDR24330.1 CCAG010010771 KA648236 AFP62865.1 KZ288416 PBC26059.1 GBYB01001152 JAG70919.1 UFQS01000037 UFQT01000037 SSW98015.1 SSX18401.1 SSW98016.1 SSX18402.1 NWSH01000178 PCG78709.1 PZC72815.1 BABH01009804 BABH01009805 GFDF01009490 JAV04594.1 KYN06498.1 KOC69919.1 KYN43601.1 APGK01053700 APGK01053701 KB741231 ENN72266.1 JXJN01011785 KYQ50179.1 CCAG010012933 KQ761655 OAD57197.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000283053
UP000007755
+ More
UP000008237 UP000078542 UP000078492 UP000005205 UP000053097 UP000279307 UP000076502 UP000075809 UP000000311 UP000053825 UP000218220 UP000007266 UP000078541 UP000268350 UP000095301 UP000007798 UP000009192 UP000001819 UP000183832 UP000037069 UP000192221 UP000008711 UP000215335 UP000000803 UP000078540 UP000002282 UP000007801 UP000015103 UP000075900 UP000009046 UP000076408 UP000075882 UP000007062 UP000075903 UP000075886 UP000075902 UP000008792 UP000092553 UP000095300 UP000235965 UP000092443 UP000078200 UP000036403 UP000027135 UP000092444 UP000091820 UP000242457 UP000092445 UP000079169 UP000005203 UP000019118 UP000092460
UP000008237 UP000078542 UP000078492 UP000005205 UP000053097 UP000279307 UP000076502 UP000075809 UP000000311 UP000053825 UP000218220 UP000007266 UP000078541 UP000268350 UP000095301 UP000007798 UP000009192 UP000001819 UP000183832 UP000037069 UP000192221 UP000008711 UP000215335 UP000000803 UP000078540 UP000002282 UP000007801 UP000015103 UP000075900 UP000009046 UP000076408 UP000075882 UP000007062 UP000075903 UP000075886 UP000075902 UP000008792 UP000092553 UP000095300 UP000235965 UP000092443 UP000078200 UP000036403 UP000027135 UP000092444 UP000091820 UP000242457 UP000092445 UP000079169 UP000005203 UP000019118 UP000092460
Interpro
Gene 3D
ProteinModelPortal
H9JNX2
A0A2W1BJ02
A0A194RU78
A0A194PTI5
A0A212FCX8
A0A3S2PHH3
+ More
A0A2W1BCV5 A0A2H1VWQ6 A0A212FD00 F4W561 E2BDY3 A0A195D0Q3 A0A151J3V4 A0A158NZC1 A0A026W645 A0A154PDD7 A0A194PS69 A0A151WQU6 E2AZ43 A0A194RQU6 E9J4S9 A0A0L7RG38 A0A2A4IX18 D6WHS6 A0A1B6KUA4 A0A1B6FLN6 A0A195FUP4 A0A1Y1LPP5 A0A3B0KFJ9 A0A1I8NGG5 B4MLM1 A0A310SF21 B4L9P9 A0A0A1XP62 A0A0R3P8K9 A0A1J1I7T3 A0A0K8USS0 A0A023F004 A0A0V0GBK0 W8AVR2 A0A0L0BT78 A0A1W4UXC6 B3NCD2 A0A232F2T3 Q9VRT4 A0A151I4F7 A0A0J9RR75 B4PJC0 A0A224XLE8 T1DJX3 A0A0P8YIG7 T1I0F0 A0A0L0C4M2 A0A1Q3G090 A0A3F2YXP2 E0VI57 A0A0P6IYS1 A0A182Y2J8 A0A182LKD6 Q7Q570 A0A182VNM0 A0A182QEZ0 A0A182TJ22 B4LDD7 A0A0M4EFT7 A0A1I8Q048 A0A0K8VFG7 A0A2J7R373 A0A1I8NXE0 A0A1A9Y9S3 A0A2J7R361 A0A1A9VMM4 A0A0J7NSN3 A0A067RU41 A0A1B0GG54 A0A1A9WFE2 T1PHJ0 A0A2A3E4H2 A0A1B0AIC1 A0A0C9QKP2 A0A336K3J5 A0A336JZ48 A0A2A4K3T5 A0A2W1BGI1 A0A1S3D083 A0A1A9ZKZ7 H9JNX1 A0A088A3S5 A0A1A9X3B2 A0A1L8DE47 A0A195D0Q4 A0A0L7RGH8 A0A195FT31 N6TV42 A0A1A9V9X7 A0A088AMR8 A0A1B0BC30 A0A151WQK2 A0A1B0F9D3 A0A310SBQ9
A0A2W1BCV5 A0A2H1VWQ6 A0A212FD00 F4W561 E2BDY3 A0A195D0Q3 A0A151J3V4 A0A158NZC1 A0A026W645 A0A154PDD7 A0A194PS69 A0A151WQU6 E2AZ43 A0A194RQU6 E9J4S9 A0A0L7RG38 A0A2A4IX18 D6WHS6 A0A1B6KUA4 A0A1B6FLN6 A0A195FUP4 A0A1Y1LPP5 A0A3B0KFJ9 A0A1I8NGG5 B4MLM1 A0A310SF21 B4L9P9 A0A0A1XP62 A0A0R3P8K9 A0A1J1I7T3 A0A0K8USS0 A0A023F004 A0A0V0GBK0 W8AVR2 A0A0L0BT78 A0A1W4UXC6 B3NCD2 A0A232F2T3 Q9VRT4 A0A151I4F7 A0A0J9RR75 B4PJC0 A0A224XLE8 T1DJX3 A0A0P8YIG7 T1I0F0 A0A0L0C4M2 A0A1Q3G090 A0A3F2YXP2 E0VI57 A0A0P6IYS1 A0A182Y2J8 A0A182LKD6 Q7Q570 A0A182VNM0 A0A182QEZ0 A0A182TJ22 B4LDD7 A0A0M4EFT7 A0A1I8Q048 A0A0K8VFG7 A0A2J7R373 A0A1I8NXE0 A0A1A9Y9S3 A0A2J7R361 A0A1A9VMM4 A0A0J7NSN3 A0A067RU41 A0A1B0GG54 A0A1A9WFE2 T1PHJ0 A0A2A3E4H2 A0A1B0AIC1 A0A0C9QKP2 A0A336K3J5 A0A336JZ48 A0A2A4K3T5 A0A2W1BGI1 A0A1S3D083 A0A1A9ZKZ7 H9JNX1 A0A088A3S5 A0A1A9X3B2 A0A1L8DE47 A0A195D0Q4 A0A0L7RGH8 A0A195FT31 N6TV42 A0A1A9V9X7 A0A088AMR8 A0A1B0BC30 A0A151WQK2 A0A1B0F9D3 A0A310SBQ9
PDB
5EQI
E-value=0.000183463,
Score=107
Ontologies
GO
Topology
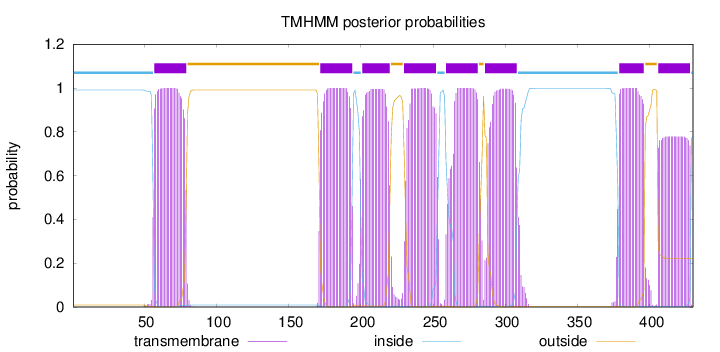
Length:
430
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
167.56237
Exp number, first 60 AAs:
4.34511
Total prob of N-in:
0.99107
inside
1 - 56
TMhelix
57 - 79
outside
80 - 171
TMhelix
172 - 194
inside
195 - 200
TMhelix
201 - 220
outside
221 - 229
TMhelix
230 - 252
inside
253 - 258
TMhelix
259 - 281
outside
282 - 285
TMhelix
286 - 308
inside
309 - 378
TMhelix
379 - 396
outside
397 - 405
TMhelix
406 - 428
inside
429 - 430
Population Genetic Test Statistics
Pi
239.881476
Theta
171.465312
Tajima's D
1.307294
CLR
0.238398
CSRT
0.739613019349033
Interpretation
Uncertain
