Gene
KWMTBOMO14065
Pre Gene Modal
BGIBMGA011226
Annotation
mfs_transporter_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 2.861
Sequence
CDS
ATGCAGTACGCAGCCGAGATTCTGCCCACCGTCGTGAGAGCTCAGGGCCTAGCTCTGATACACATCACTGGATATGTTGCAACAATACTGGTGCCGTACATCGTATATTTGGCAACCATTGCGACCGAAATACCCCTTCTAATATTAGGCACTGTGGGCATAATTGGTGGAATTTTATGCCTCTTTCTACCGGAATCGATGGGCAAAGAAATGCCTCAAACTCTTCAGGATGGAGAAGAGTATGGGTTGGAGCAGAAGTTCTGGGATTTTCCGTGCTGCACCAGGTGGGTTAGCCAAGACTCGGGTCTGCTATCATAG
Protein
MQYAAEILPTVVRAQGLALIHITGYVATILVPYIVYLATIATEIPLLILGTVGIIGGILCLFLPESMGKEMPQTLQDGEEYGLEQKFWDFPCCTRWVSQDSGLLS
Summary
Uniprot
H9JNX2
A0A2H1VWQ6
I4DQ48
A0A3S2PHH3
A0A194RU78
A0A212FCX8
+ More
A0A194PTI5 A0A2W1BJ02 A0A0T6BGS7 D6WHS6 A0A1B6CJD4 A0A2J7R361 A0A3Q0IY51 H9JNX1 A0A194PS69 A0A194RQU6 A0A2A4K3T5 A0A182SFW5 A0A1J1I7T3 A0A067RU41 U4UVU8 A0A1B0GPP7 A0A1B0CQJ9 A0A1B6IA56 A0A088A3S5 A0A2W1BGI1 A0A1Y1LPP5 N6TV42 Q16MV3 A0A1L8DE47 A0A2H1WBW6 A0A154PDD7 A0A182H633 A0A0P6IYS1 Q16K11 A0A224XLE8 A0A2A3E4H2 A0A0V0GBK0 A0A0L0C4M2 A0A023F004 T1I0F0 A0A0J7KEI0 E0VI57 A0A336JZ48 A0A336K3J5 A0A1B6KUA4 A0A182GGU3 A0A182NI96 A0A034VS57 A0A0L7RG38 E2BDY3 A0A182JGG5 B0XLV9 A0A182JYN2 A0A1B6FLN6 K7J144 A0A182Y2J8 A0A182I4I9 A0A182X4J2 Q7Q570 A0A182LKD6 B0XGJ2 A0A182TJ22 A0A182VNM0 A0A3F2YXP2 A0A182WBQ6 A0A182FGA1 A0A310SF21 A0A1A9WFE2 A0A182PMU9 A0A232F2T3 T1DJX3 A0A182QEZ0 W5J8J5 A0A0M4EFT7 A0A1S3D083 B4IZZ0 A0A182M546 A0A1Q3EVR9 W8AZU8 A0A034VTY6 A0A084WGJ3 A0A2R7WAA3 A0A1B0AIC1 A0A1Q3G090 A0A0L0BT78 W8AVR2 T1PHJ0 V9IIX1 A0A1A9Y9S3 A0A1A9VMM4 A0A1B0GG54 A0A026W645 A0A0P5KEP7 A0A336MA72 A0A0A1XP62 Q2LZD0 A0A3B0KFJ9 A0A1I8NGG5 A0A0R3P8K9
A0A194PTI5 A0A2W1BJ02 A0A0T6BGS7 D6WHS6 A0A1B6CJD4 A0A2J7R361 A0A3Q0IY51 H9JNX1 A0A194PS69 A0A194RQU6 A0A2A4K3T5 A0A182SFW5 A0A1J1I7T3 A0A067RU41 U4UVU8 A0A1B0GPP7 A0A1B0CQJ9 A0A1B6IA56 A0A088A3S5 A0A2W1BGI1 A0A1Y1LPP5 N6TV42 Q16MV3 A0A1L8DE47 A0A2H1WBW6 A0A154PDD7 A0A182H633 A0A0P6IYS1 Q16K11 A0A224XLE8 A0A2A3E4H2 A0A0V0GBK0 A0A0L0C4M2 A0A023F004 T1I0F0 A0A0J7KEI0 E0VI57 A0A336JZ48 A0A336K3J5 A0A1B6KUA4 A0A182GGU3 A0A182NI96 A0A034VS57 A0A0L7RG38 E2BDY3 A0A182JGG5 B0XLV9 A0A182JYN2 A0A1B6FLN6 K7J144 A0A182Y2J8 A0A182I4I9 A0A182X4J2 Q7Q570 A0A182LKD6 B0XGJ2 A0A182TJ22 A0A182VNM0 A0A3F2YXP2 A0A182WBQ6 A0A182FGA1 A0A310SF21 A0A1A9WFE2 A0A182PMU9 A0A232F2T3 T1DJX3 A0A182QEZ0 W5J8J5 A0A0M4EFT7 A0A1S3D083 B4IZZ0 A0A182M546 A0A1Q3EVR9 W8AZU8 A0A034VTY6 A0A084WGJ3 A0A2R7WAA3 A0A1B0AIC1 A0A1Q3G090 A0A0L0BT78 W8AVR2 T1PHJ0 V9IIX1 A0A1A9Y9S3 A0A1A9VMM4 A0A1B0GG54 A0A026W645 A0A0P5KEP7 A0A336MA72 A0A0A1XP62 Q2LZD0 A0A3B0KFJ9 A0A1I8NGG5 A0A0R3P8K9
Pubmed
19121390
22651552
26354079
22118469
28756777
18362917
+ More
19820115 24845553 23537049 28004739 17510324 26483478 26999592 26108605 25474469 20566863 25348373 20798317 20075255 25244985 12364791 14747013 17210077 20966253 28648823 20920257 23761445 17994087 24495485 24438588 25315136 24508170 30249741 25830018 15632085
19820115 24845553 23537049 28004739 17510324 26483478 26999592 26108605 25474469 20566863 25348373 20798317 20075255 25244985 12364791 14747013 17210077 20966253 28648823 20920257 23761445 17994087 24495485 24438588 25315136 24508170 30249741 25830018 15632085
EMBL
BABH01009800
BABH01009801
ODYU01004909
SOQ45269.1
AK403928
BAM20038.1
+ More
RSAL01000031 RVE51608.1 KQ459875 KPJ19681.1 AGBW02009123 OWR51601.1 KQ459593 KPI96288.1 KZ150153 PZC72816.1 LJIG01000424 KRT86516.1 KQ971321 EFA00687.1 GEDC01023768 JAS13530.1 NEVH01007822 PNF35272.1 BABH01009804 BABH01009805 KPI96286.1 KPJ19680.1 NWSH01000178 PCG78709.1 CVRI01000042 CRK95636.1 KK852417 KDR24330.1 KB632387 ERL94366.1 AJVK01015236 AJWK01023693 GECU01023907 JAS83799.1 PZC72815.1 GEZM01052731 JAV74290.1 APGK01053700 APGK01053701 KB741231 ENN72266.1 CH477848 EAT35660.1 GFDF01009490 JAV04594.1 ODYU01007620 SOQ50550.1 KQ434878 KZC09853.1 JXUM01026950 JXUM01026951 JXUM01026952 JXUM01026953 KQ560772 KXJ80949.1 GDUN01000921 JAN94998.1 CH477981 EAT34641.1 GFTR01007453 JAW08973.1 KZ288416 PBC26059.1 GECL01001415 JAP04709.1 JRES01000930 KNC27191.1 GBBI01004623 JAC14089.1 ACPB03009635 ACPB03009636 ACPB03009637 ACPB03009638 ACPB03009639 LBMM01008789 KMQ88639.1 DS235184 EEB13063.1 UFQS01000037 UFQT01000037 SSW98016.1 SSX18402.1 SSW98015.1 SSX18401.1 GEBQ01024996 JAT14981.1 JXUM01062362 KQ562196 KXJ76434.1 GAKP01013673 JAC45279.1 KQ414596 KOC69917.1 GL447693 EFN86109.1 DS234836 EDS35332.1 GECZ01018651 JAS51118.1 APCN01002315 AAAB01008960 EAA11678.4 DS233035 EDS27656.1 KQ769818 OAD52819.1 NNAY01001101 OXU25136.1 GAMD01001231 JAB00360.1 AXCN02000878 ADMH02001954 ETN60286.1 CP012525 ALC43177.1 CH916366 EDV96762.1 AXCM01001383 GFDL01015640 JAV19405.1 GAMC01016162 JAB90393.1 GAKP01013667 JAC45285.1 ATLV01023587 KE525344 KFB49337.1 KK854453 PTY15970.1 GFDL01001826 JAV33219.1 JRES01001371 KNC23290.1 GAMC01016163 JAB90392.1 KA648236 AFP62865.1 JR047570 AEY60446.1 CCAG010010771 KK107419 QOIP01000003 EZA51081.1 RLU24986.1 GDIQ01186847 JAK64878.1 UFQT01000295 SSX22938.1 GBXI01001560 JAD12732.1 CH379069 EAL29580.3 OUUW01000009 SPP85099.1 KRT08150.1
RSAL01000031 RVE51608.1 KQ459875 KPJ19681.1 AGBW02009123 OWR51601.1 KQ459593 KPI96288.1 KZ150153 PZC72816.1 LJIG01000424 KRT86516.1 KQ971321 EFA00687.1 GEDC01023768 JAS13530.1 NEVH01007822 PNF35272.1 BABH01009804 BABH01009805 KPI96286.1 KPJ19680.1 NWSH01000178 PCG78709.1 CVRI01000042 CRK95636.1 KK852417 KDR24330.1 KB632387 ERL94366.1 AJVK01015236 AJWK01023693 GECU01023907 JAS83799.1 PZC72815.1 GEZM01052731 JAV74290.1 APGK01053700 APGK01053701 KB741231 ENN72266.1 CH477848 EAT35660.1 GFDF01009490 JAV04594.1 ODYU01007620 SOQ50550.1 KQ434878 KZC09853.1 JXUM01026950 JXUM01026951 JXUM01026952 JXUM01026953 KQ560772 KXJ80949.1 GDUN01000921 JAN94998.1 CH477981 EAT34641.1 GFTR01007453 JAW08973.1 KZ288416 PBC26059.1 GECL01001415 JAP04709.1 JRES01000930 KNC27191.1 GBBI01004623 JAC14089.1 ACPB03009635 ACPB03009636 ACPB03009637 ACPB03009638 ACPB03009639 LBMM01008789 KMQ88639.1 DS235184 EEB13063.1 UFQS01000037 UFQT01000037 SSW98016.1 SSX18402.1 SSW98015.1 SSX18401.1 GEBQ01024996 JAT14981.1 JXUM01062362 KQ562196 KXJ76434.1 GAKP01013673 JAC45279.1 KQ414596 KOC69917.1 GL447693 EFN86109.1 DS234836 EDS35332.1 GECZ01018651 JAS51118.1 APCN01002315 AAAB01008960 EAA11678.4 DS233035 EDS27656.1 KQ769818 OAD52819.1 NNAY01001101 OXU25136.1 GAMD01001231 JAB00360.1 AXCN02000878 ADMH02001954 ETN60286.1 CP012525 ALC43177.1 CH916366 EDV96762.1 AXCM01001383 GFDL01015640 JAV19405.1 GAMC01016162 JAB90393.1 GAKP01013667 JAC45285.1 ATLV01023587 KE525344 KFB49337.1 KK854453 PTY15970.1 GFDL01001826 JAV33219.1 JRES01001371 KNC23290.1 GAMC01016163 JAB90392.1 KA648236 AFP62865.1 JR047570 AEY60446.1 CCAG010010771 KK107419 QOIP01000003 EZA51081.1 RLU24986.1 GDIQ01186847 JAK64878.1 UFQT01000295 SSX22938.1 GBXI01001560 JAD12732.1 CH379069 EAL29580.3 OUUW01000009 SPP85099.1 KRT08150.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000007151
UP000053268
UP000007266
+ More
UP000235965 UP000079169 UP000218220 UP000075901 UP000183832 UP000027135 UP000030742 UP000092462 UP000092461 UP000005203 UP000019118 UP000008820 UP000076502 UP000069940 UP000249989 UP000242457 UP000037069 UP000015103 UP000036403 UP000009046 UP000075884 UP000053825 UP000008237 UP000075880 UP000002320 UP000075881 UP000002358 UP000076408 UP000075840 UP000076407 UP000007062 UP000075882 UP000075902 UP000075903 UP000075900 UP000075920 UP000069272 UP000091820 UP000075885 UP000215335 UP000075886 UP000000673 UP000092553 UP000001070 UP000075883 UP000030765 UP000092445 UP000095301 UP000092443 UP000078200 UP000092444 UP000053097 UP000279307 UP000001819 UP000268350
UP000235965 UP000079169 UP000218220 UP000075901 UP000183832 UP000027135 UP000030742 UP000092462 UP000092461 UP000005203 UP000019118 UP000008820 UP000076502 UP000069940 UP000249989 UP000242457 UP000037069 UP000015103 UP000036403 UP000009046 UP000075884 UP000053825 UP000008237 UP000075880 UP000002320 UP000075881 UP000002358 UP000076408 UP000075840 UP000076407 UP000007062 UP000075882 UP000075902 UP000075903 UP000075900 UP000075920 UP000069272 UP000091820 UP000075885 UP000215335 UP000075886 UP000000673 UP000092553 UP000001070 UP000075883 UP000030765 UP000092445 UP000095301 UP000092443 UP000078200 UP000092444 UP000053097 UP000279307 UP000001819 UP000268350
Interpro
SUPFAM
SSF103473
SSF103473
ProteinModelPortal
H9JNX2
A0A2H1VWQ6
I4DQ48
A0A3S2PHH3
A0A194RU78
A0A212FCX8
+ More
A0A194PTI5 A0A2W1BJ02 A0A0T6BGS7 D6WHS6 A0A1B6CJD4 A0A2J7R361 A0A3Q0IY51 H9JNX1 A0A194PS69 A0A194RQU6 A0A2A4K3T5 A0A182SFW5 A0A1J1I7T3 A0A067RU41 U4UVU8 A0A1B0GPP7 A0A1B0CQJ9 A0A1B6IA56 A0A088A3S5 A0A2W1BGI1 A0A1Y1LPP5 N6TV42 Q16MV3 A0A1L8DE47 A0A2H1WBW6 A0A154PDD7 A0A182H633 A0A0P6IYS1 Q16K11 A0A224XLE8 A0A2A3E4H2 A0A0V0GBK0 A0A0L0C4M2 A0A023F004 T1I0F0 A0A0J7KEI0 E0VI57 A0A336JZ48 A0A336K3J5 A0A1B6KUA4 A0A182GGU3 A0A182NI96 A0A034VS57 A0A0L7RG38 E2BDY3 A0A182JGG5 B0XLV9 A0A182JYN2 A0A1B6FLN6 K7J144 A0A182Y2J8 A0A182I4I9 A0A182X4J2 Q7Q570 A0A182LKD6 B0XGJ2 A0A182TJ22 A0A182VNM0 A0A3F2YXP2 A0A182WBQ6 A0A182FGA1 A0A310SF21 A0A1A9WFE2 A0A182PMU9 A0A232F2T3 T1DJX3 A0A182QEZ0 W5J8J5 A0A0M4EFT7 A0A1S3D083 B4IZZ0 A0A182M546 A0A1Q3EVR9 W8AZU8 A0A034VTY6 A0A084WGJ3 A0A2R7WAA3 A0A1B0AIC1 A0A1Q3G090 A0A0L0BT78 W8AVR2 T1PHJ0 V9IIX1 A0A1A9Y9S3 A0A1A9VMM4 A0A1B0GG54 A0A026W645 A0A0P5KEP7 A0A336MA72 A0A0A1XP62 Q2LZD0 A0A3B0KFJ9 A0A1I8NGG5 A0A0R3P8K9
A0A194PTI5 A0A2W1BJ02 A0A0T6BGS7 D6WHS6 A0A1B6CJD4 A0A2J7R361 A0A3Q0IY51 H9JNX1 A0A194PS69 A0A194RQU6 A0A2A4K3T5 A0A182SFW5 A0A1J1I7T3 A0A067RU41 U4UVU8 A0A1B0GPP7 A0A1B0CQJ9 A0A1B6IA56 A0A088A3S5 A0A2W1BGI1 A0A1Y1LPP5 N6TV42 Q16MV3 A0A1L8DE47 A0A2H1WBW6 A0A154PDD7 A0A182H633 A0A0P6IYS1 Q16K11 A0A224XLE8 A0A2A3E4H2 A0A0V0GBK0 A0A0L0C4M2 A0A023F004 T1I0F0 A0A0J7KEI0 E0VI57 A0A336JZ48 A0A336K3J5 A0A1B6KUA4 A0A182GGU3 A0A182NI96 A0A034VS57 A0A0L7RG38 E2BDY3 A0A182JGG5 B0XLV9 A0A182JYN2 A0A1B6FLN6 K7J144 A0A182Y2J8 A0A182I4I9 A0A182X4J2 Q7Q570 A0A182LKD6 B0XGJ2 A0A182TJ22 A0A182VNM0 A0A3F2YXP2 A0A182WBQ6 A0A182FGA1 A0A310SF21 A0A1A9WFE2 A0A182PMU9 A0A232F2T3 T1DJX3 A0A182QEZ0 W5J8J5 A0A0M4EFT7 A0A1S3D083 B4IZZ0 A0A182M546 A0A1Q3EVR9 W8AZU8 A0A034VTY6 A0A084WGJ3 A0A2R7WAA3 A0A1B0AIC1 A0A1Q3G090 A0A0L0BT78 W8AVR2 T1PHJ0 V9IIX1 A0A1A9Y9S3 A0A1A9VMM4 A0A1B0GG54 A0A026W645 A0A0P5KEP7 A0A336MA72 A0A0A1XP62 Q2LZD0 A0A3B0KFJ9 A0A1I8NGG5 A0A0R3P8K9
Ontologies
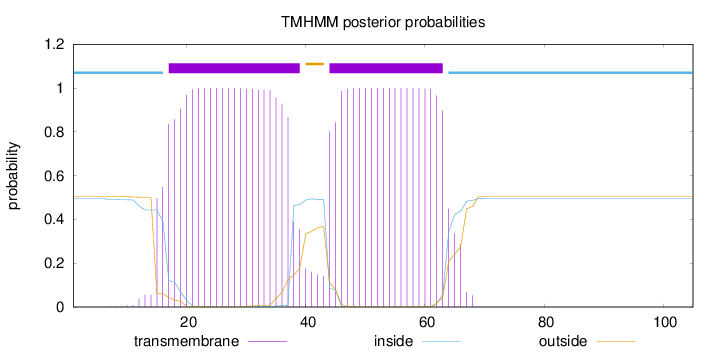
Topology
Length:
105
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.53148
Exp number, first 60 AAs:
39.47587
Total prob of N-in:
0.49530
POSSIBLE N-term signal
sequence
inside
1 - 16
TMhelix
17 - 39
outside
40 - 43
TMhelix
44 - 63
inside
64 - 105
Population Genetic Test Statistics
Pi
286.344285
Theta
144.739838
Tajima's D
3.158828
CLR
0.118604
CSRT
0.985250737463127
Interpretation
Uncertain
