Gene
KWMTBOMO14059
Pre Gene Modal
BGIBMGA011229
Annotation
PREDICTED:_NADH_dehydrogenase_[ubiquinone]_1_alpha_subcomplex_assembly_factor_3_[Papilio_polytes]
Full name
DNA polymerase
+ More
S-adenosylmethionine sensor upstream of mTORC1
S-adenosylmethionine sensor upstream of mTORC1
Alternative Name
Probable methyltransferase BMT2 homolog
Location in the cell
Cytoplasmic Reliability : 1.125 Mitochondrial Reliability : 1.323
Sequence
CDS
ATGTTTCAAAATACAATAAGGCATATCAACGTTTTATCAAAAAGAAAACTTCCTATTTTGAGTTTTACACGTACCAAAGCAGCGTACGAAGGAGAGGGCAAGACGACTGTGAGAGTTCTGAACAACGAATATGACCTCGGACTAATGATTGACTCTTTTGCTACTTATGGCTTCAAGTTAAACAACGGCATCACAGTTTTAGGACCTATGGCCATATTTCCAAGAACGGTATTATCTTGGCAAGTCGCAGATTCGGATGACGTCACCGAGGAGTCCCTGAAGCTGTTCAAACTGCTCGAGCCTAGAATCGATCTGCTTATCTTAGGCTTGGATACGACGGATCGTTCGAAAATAGACAAAGCATTCAGGGCGGCCCGCGCGAACGAATTGAACATCGAAATCTTGGCAACAGAACACGCATGTTCCACATTCAACTTCCTGAACGCAGAGGGCAGATCCGTGGCTGCTGCCCTGATACCACCGTTCAAAGTCAACATGAACGAAGACGATATGCTTGAAGTGAAGATGCACTACAGTGATATATTCCAGAAAGGATTCTCGAAGCCTTCAGGGAAATAA
Protein
MFQNTIRHINVLSKRKLPILSFTRTKAAYEGEGKTTVRVLNNEYDLGLMIDSFATYGFKLNNGITVLGPMAIFPRTVLSWQVADSDDVTEESLKLFKLLEPRIDLLILGLDTTDRSKIDKAFRAARANELNIEILATEHACSTFNFLNAEGRSVAAALIPPFKVNMNEDDMLEVKMHYSDIFQKGFSKPSGK
Summary
Description
S-adenosyl-L-methionine-binding protein that acts as an inhibitor of mTORC1 signaling. Acts as a sensor of S-adenosyl-L-methionine to signal methionine sufficiency to mTORC1. Probably also acts as a S-adenosyl-L-methionine-dependent methyltransferase.
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Cofactor
[4Fe-4S] cluster
Similarity
Belongs to the DNA polymerase type-B family.
Belongs to the BMT2 family.
Belongs to the BMT2 family.
Uniprot
A0A194PYW2
A0A194RIX3
A0A2A4JHQ1
A0A2W1BPT6
A0A212FD82
A0A2H1VTP6
+ More
A0A182GMI0 A0A1Q3FGE1 J3JXA2 B0WKL7 B0XLX7 A0A0K8URV5 A0A0L7L739 Q7QFV4 A0A182UCW5 A0A182VM34 A0A182LAP0 A0A182WV37 A0A182I8R1 A0A0A1XFM0 Q16XR7 U5EV21 A0A1Y1NCV0 A0A182P765 A0A084VC13 A0A182VXT2 A0A182NFV5 A0A182XXC0 A0A182JNQ3 B4KMD7 B4LNX6 A0A182Q9R6 A0A224XLX3 A0A182RQP5 A0A182JFZ4 W5JHC1 A0A2M4AY75 B4MRP9 A0A1D2NGN4 A0A2P8YKV0 A0A2M4C1G6 A0A069DPU6 A0A0V0G5Q0 A0A0A9W973 A0A1L8DWL4 B3MCL0 D6WK37 A0A1B0G3M5 Q292R7 B4GCX2 A0A0K8TSG8 A0A1L8DWU0 A0A1L8DWP0 A0A0P4VML8 B4J634 A0A1A9UDK1 A0A1W4UME1 A0A1B0GLC6 A0A3B0IYT7 A0A067R8Z8 B4I8U8 A0A0M4EIJ7 A0A3B0IYJ6 Q9W1H9 B3NPU0 A0A0L0CQQ2 A0A2M3ZDG3 B4QB10 B4PA84 A0A336MSR5 A0A1B0D502 A0A1B0BCM8 A0A1A9Y7Z2 A0A087U8W1 A0A1I8Q109 A0A1L8ECH8 A0A1B6CZ84 A0A1I8N0K6 A0A1L8E986 A0A2R5LDU3 A0A293M8Z8 E9H068 A0A2L2YEI8 A0A0P5PVI4 A0A0P6JLX5 A0A1J1IBL5 A0A232F1C2 A0A131YWF7 A0A162NRU7 A0A2R7VTT7 A0A0P4VXY1 A0A0P5YC68 A0A1A9WW31 A0A1E1WW09 A0A131XDA2 A0A1B6MFV0 A0A023G5R3 A0A224YUJ3
A0A182GMI0 A0A1Q3FGE1 J3JXA2 B0WKL7 B0XLX7 A0A0K8URV5 A0A0L7L739 Q7QFV4 A0A182UCW5 A0A182VM34 A0A182LAP0 A0A182WV37 A0A182I8R1 A0A0A1XFM0 Q16XR7 U5EV21 A0A1Y1NCV0 A0A182P765 A0A084VC13 A0A182VXT2 A0A182NFV5 A0A182XXC0 A0A182JNQ3 B4KMD7 B4LNX6 A0A182Q9R6 A0A224XLX3 A0A182RQP5 A0A182JFZ4 W5JHC1 A0A2M4AY75 B4MRP9 A0A1D2NGN4 A0A2P8YKV0 A0A2M4C1G6 A0A069DPU6 A0A0V0G5Q0 A0A0A9W973 A0A1L8DWL4 B3MCL0 D6WK37 A0A1B0G3M5 Q292R7 B4GCX2 A0A0K8TSG8 A0A1L8DWU0 A0A1L8DWP0 A0A0P4VML8 B4J634 A0A1A9UDK1 A0A1W4UME1 A0A1B0GLC6 A0A3B0IYT7 A0A067R8Z8 B4I8U8 A0A0M4EIJ7 A0A3B0IYJ6 Q9W1H9 B3NPU0 A0A0L0CQQ2 A0A2M3ZDG3 B4QB10 B4PA84 A0A336MSR5 A0A1B0D502 A0A1B0BCM8 A0A1A9Y7Z2 A0A087U8W1 A0A1I8Q109 A0A1L8ECH8 A0A1B6CZ84 A0A1I8N0K6 A0A1L8E986 A0A2R5LDU3 A0A293M8Z8 E9H068 A0A2L2YEI8 A0A0P5PVI4 A0A0P6JLX5 A0A1J1IBL5 A0A232F1C2 A0A131YWF7 A0A162NRU7 A0A2R7VTT7 A0A0P4VXY1 A0A0P5YC68 A0A1A9WW31 A0A1E1WW09 A0A131XDA2 A0A1B6MFV0 A0A023G5R3 A0A224YUJ3
EC Number
2.7.7.7
Pubmed
26354079
28756777
22118469
26483478
22516182
23537049
+ More
26227816 12364791 14747013 17210077 20966253 25830018 17510324 28004739 24438588 25244985 17994087 20920257 23761445 27289101 29403074 26334808 25401762 26823975 18362917 19820115 15632085 26369729 27129103 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 22936249 17550304 25315136 21292972 26561354 28648823 26830274 28503490 28049606 28797301
26227816 12364791 14747013 17210077 20966253 25830018 17510324 28004739 24438588 25244985 17994087 20920257 23761445 27289101 29403074 26334808 25401762 26823975 18362917 19820115 15632085 26369729 27129103 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 22936249 17550304 25315136 21292972 26561354 28648823 26830274 28503490 28049606 28797301
EMBL
KQ459593
KPI96345.1
KQ460152
KPJ17389.1
NWSH01001410
PCG71339.1
+ More
KZ149962 PZC76301.1 AGBW02009108 OWR51658.1 ODYU01004375 SOQ44190.1 JXUM01074654 KQ562843 KXJ75028.1 GFDL01008426 JAV26619.1 APGK01032733 BT127871 KB740848 KB631797 AEE62833.1 ENN78710.1 ERL86209.1 DS231974 EDS29948.1 DS234896 EDS35440.1 GDHF01022887 JAI29427.1 JTDY01002591 KOB71144.1 AAAB01008844 EAA05972.2 APCN01003108 GBXI01004556 JAD09736.1 CH477534 EAT39375.1 GANO01003606 JAB56265.1 GEZM01011790 JAV93437.1 ATLV01009976 KE524560 KFB35507.1 CH933808 EDW09825.1 CH940648 EDW61145.1 AXCN02000249 GFTR01002940 JAW13486.1 ADMH02001495 ETN62315.1 GGFK01012410 MBW45731.1 CH963850 EDW74788.1 LJIJ01000043 ODN04420.1 PYGN01000522 PSN44879.1 GGFJ01009687 MBW58828.1 GBGD01003178 JAC85711.1 GECL01002713 JAP03411.1 GBHO01042204 GBHO01029750 GBRD01003389 GDHC01008226 JAG01400.1 JAG13854.1 JAG62432.1 JAQ10403.1 GFDF01003243 JAV10841.1 CH902619 EDV36244.1 KQ971342 EFA04623.2 CCAG010010932 CM000071 EAL24794.1 CH479181 EDW31510.1 GDAI01000296 JAI17307.1 GFDF01003198 JAV10886.1 GFDF01003242 JAV10842.1 GDKW01002505 JAI54090.1 CH916367 EDW01892.1 AJWK01034746 OUUW01000001 SPP73155.1 KK852618 KDR20135.1 CH480824 EDW57023.1 CP012524 ALC42610.1 SPP73154.1 AE013599 AY119235 AAF47082.1 AAM51095.1 CH954179 EDV56881.1 JRES01000153 KNC33764.1 GGFM01005835 MBW26586.1 CM000362 CM002911 EDX08447.1 KMY96141.1 CM000158 EDW92410.1 UFQT01002082 SSX32601.1 AJVK01025011 JXJN01012064 KK118761 KFM73800.1 GFDG01002378 JAV16421.1 GEDC01018519 JAS18779.1 GFDG01003630 JAV15169.1 GGLE01003381 MBY07507.1 GFWV01012087 MAA36816.1 GL732580 EFX74747.1 IAAA01028667 IAAA01028668 LAA06548.1 GDIQ01123606 JAL28120.1 GDIQ01014558 JAN80179.1 CVRI01000047 CRK97603.1 NNAY01001262 OXU24576.1 GEDV01005712 JAP82845.1 LRGB01000547 KZS18213.1 KK854085 PTY10944.1 GDRN01099349 JAI58780.1 GDIP01059772 JAM43943.1 GFAC01007975 JAT91213.1 GEFH01005020 JAP63561.1 GEBQ01010098 GEBQ01005202 JAT29879.1 JAT34775.1 GBBM01006279 JAC29139.1 GFPF01010122 MAA21268.1
KZ149962 PZC76301.1 AGBW02009108 OWR51658.1 ODYU01004375 SOQ44190.1 JXUM01074654 KQ562843 KXJ75028.1 GFDL01008426 JAV26619.1 APGK01032733 BT127871 KB740848 KB631797 AEE62833.1 ENN78710.1 ERL86209.1 DS231974 EDS29948.1 DS234896 EDS35440.1 GDHF01022887 JAI29427.1 JTDY01002591 KOB71144.1 AAAB01008844 EAA05972.2 APCN01003108 GBXI01004556 JAD09736.1 CH477534 EAT39375.1 GANO01003606 JAB56265.1 GEZM01011790 JAV93437.1 ATLV01009976 KE524560 KFB35507.1 CH933808 EDW09825.1 CH940648 EDW61145.1 AXCN02000249 GFTR01002940 JAW13486.1 ADMH02001495 ETN62315.1 GGFK01012410 MBW45731.1 CH963850 EDW74788.1 LJIJ01000043 ODN04420.1 PYGN01000522 PSN44879.1 GGFJ01009687 MBW58828.1 GBGD01003178 JAC85711.1 GECL01002713 JAP03411.1 GBHO01042204 GBHO01029750 GBRD01003389 GDHC01008226 JAG01400.1 JAG13854.1 JAG62432.1 JAQ10403.1 GFDF01003243 JAV10841.1 CH902619 EDV36244.1 KQ971342 EFA04623.2 CCAG010010932 CM000071 EAL24794.1 CH479181 EDW31510.1 GDAI01000296 JAI17307.1 GFDF01003198 JAV10886.1 GFDF01003242 JAV10842.1 GDKW01002505 JAI54090.1 CH916367 EDW01892.1 AJWK01034746 OUUW01000001 SPP73155.1 KK852618 KDR20135.1 CH480824 EDW57023.1 CP012524 ALC42610.1 SPP73154.1 AE013599 AY119235 AAF47082.1 AAM51095.1 CH954179 EDV56881.1 JRES01000153 KNC33764.1 GGFM01005835 MBW26586.1 CM000362 CM002911 EDX08447.1 KMY96141.1 CM000158 EDW92410.1 UFQT01002082 SSX32601.1 AJVK01025011 JXJN01012064 KK118761 KFM73800.1 GFDG01002378 JAV16421.1 GEDC01018519 JAS18779.1 GFDG01003630 JAV15169.1 GGLE01003381 MBY07507.1 GFWV01012087 MAA36816.1 GL732580 EFX74747.1 IAAA01028667 IAAA01028668 LAA06548.1 GDIQ01123606 JAL28120.1 GDIQ01014558 JAN80179.1 CVRI01000047 CRK97603.1 NNAY01001262 OXU24576.1 GEDV01005712 JAP82845.1 LRGB01000547 KZS18213.1 KK854085 PTY10944.1 GDRN01099349 JAI58780.1 GDIP01059772 JAM43943.1 GFAC01007975 JAT91213.1 GEFH01005020 JAP63561.1 GEBQ01010098 GEBQ01005202 JAT29879.1 JAT34775.1 GBBM01006279 JAC29139.1 GFPF01010122 MAA21268.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000007151
UP000069940
UP000249989
+ More
UP000019118 UP000030742 UP000002320 UP000037510 UP000007062 UP000075902 UP000075903 UP000075882 UP000076407 UP000075840 UP000008820 UP000075885 UP000030765 UP000075920 UP000075884 UP000076408 UP000075881 UP000009192 UP000008792 UP000075886 UP000075900 UP000075880 UP000000673 UP000007798 UP000094527 UP000245037 UP000007801 UP000007266 UP000092444 UP000001819 UP000008744 UP000001070 UP000078200 UP000192221 UP000092461 UP000268350 UP000027135 UP000001292 UP000092553 UP000000803 UP000008711 UP000037069 UP000000304 UP000002282 UP000092462 UP000092460 UP000092443 UP000054359 UP000095300 UP000095301 UP000000305 UP000183832 UP000215335 UP000076858 UP000091820
UP000019118 UP000030742 UP000002320 UP000037510 UP000007062 UP000075902 UP000075903 UP000075882 UP000076407 UP000075840 UP000008820 UP000075885 UP000030765 UP000075920 UP000075884 UP000076408 UP000075881 UP000009192 UP000008792 UP000075886 UP000075900 UP000075880 UP000000673 UP000007798 UP000094527 UP000245037 UP000007801 UP000007266 UP000092444 UP000001819 UP000008744 UP000001070 UP000078200 UP000192221 UP000092461 UP000268350 UP000027135 UP000001292 UP000092553 UP000000803 UP000008711 UP000037069 UP000000304 UP000002282 UP000092462 UP000092460 UP000092443 UP000054359 UP000095300 UP000095301 UP000000305 UP000183832 UP000215335 UP000076858 UP000091820
PRIDE
Pfam
Interpro
IPR036748
MTH938-like_sf
+ More
IPR007523 NDUFAF3/AAMDC
IPR034095 NDUF3
IPR042087 DNA_pol_B_C
IPR023211 DNA_pol_palm_dom_sf
IPR015088 Znf_DNA-dir_DNA_pol_B_alpha
IPR012337 RNaseH-like_sf
IPR017964 DNA-dir_DNA_pol_B_CS
IPR006134 DNA-dir_DNA_pol_B_multi_dom
IPR038256 Pol_alpha_znc_sf
IPR024647 DNA_pol_a_cat_su_N
IPR006133 DNA-dir_DNA_pol_B_exonuc
IPR036397 RNaseH_sf
IPR006172 DNA-dir_DNA_pol_B
IPR041973 KOW_Spt5_1
IPR022581 Spt5_N
IPR041975 KOW_Spt5_2
IPR041978 KOW_Spt5_5
IPR021867 Bmt2/SAMTOR
IPR005100 NGN-domain
IPR041977 KOW_Spt5_4
IPR014722 Rib_L2_dom2
IPR029063 SAM-dependent_MTases
IPR005824 KOW
IPR039385 NGN_Euk
IPR036735 NGN_dom_sf
IPR024945 Spt5_C_dom
IPR006645 NGN_dom
IPR041976 KOW_Spt5_3
IPR008991 Translation_prot_SH3-like_sf
IPR039659 SPT5
IPR007523 NDUFAF3/AAMDC
IPR034095 NDUF3
IPR042087 DNA_pol_B_C
IPR023211 DNA_pol_palm_dom_sf
IPR015088 Znf_DNA-dir_DNA_pol_B_alpha
IPR012337 RNaseH-like_sf
IPR017964 DNA-dir_DNA_pol_B_CS
IPR006134 DNA-dir_DNA_pol_B_multi_dom
IPR038256 Pol_alpha_znc_sf
IPR024647 DNA_pol_a_cat_su_N
IPR006133 DNA-dir_DNA_pol_B_exonuc
IPR036397 RNaseH_sf
IPR006172 DNA-dir_DNA_pol_B
IPR041973 KOW_Spt5_1
IPR022581 Spt5_N
IPR041975 KOW_Spt5_2
IPR041978 KOW_Spt5_5
IPR021867 Bmt2/SAMTOR
IPR005100 NGN-domain
IPR041977 KOW_Spt5_4
IPR014722 Rib_L2_dom2
IPR029063 SAM-dependent_MTases
IPR005824 KOW
IPR039385 NGN_Euk
IPR036735 NGN_dom_sf
IPR024945 Spt5_C_dom
IPR006645 NGN_dom
IPR041976 KOW_Spt5_3
IPR008991 Translation_prot_SH3-like_sf
IPR039659 SPT5
CDD
ProteinModelPortal
A0A194PYW2
A0A194RIX3
A0A2A4JHQ1
A0A2W1BPT6
A0A212FD82
A0A2H1VTP6
+ More
A0A182GMI0 A0A1Q3FGE1 J3JXA2 B0WKL7 B0XLX7 A0A0K8URV5 A0A0L7L739 Q7QFV4 A0A182UCW5 A0A182VM34 A0A182LAP0 A0A182WV37 A0A182I8R1 A0A0A1XFM0 Q16XR7 U5EV21 A0A1Y1NCV0 A0A182P765 A0A084VC13 A0A182VXT2 A0A182NFV5 A0A182XXC0 A0A182JNQ3 B4KMD7 B4LNX6 A0A182Q9R6 A0A224XLX3 A0A182RQP5 A0A182JFZ4 W5JHC1 A0A2M4AY75 B4MRP9 A0A1D2NGN4 A0A2P8YKV0 A0A2M4C1G6 A0A069DPU6 A0A0V0G5Q0 A0A0A9W973 A0A1L8DWL4 B3MCL0 D6WK37 A0A1B0G3M5 Q292R7 B4GCX2 A0A0K8TSG8 A0A1L8DWU0 A0A1L8DWP0 A0A0P4VML8 B4J634 A0A1A9UDK1 A0A1W4UME1 A0A1B0GLC6 A0A3B0IYT7 A0A067R8Z8 B4I8U8 A0A0M4EIJ7 A0A3B0IYJ6 Q9W1H9 B3NPU0 A0A0L0CQQ2 A0A2M3ZDG3 B4QB10 B4PA84 A0A336MSR5 A0A1B0D502 A0A1B0BCM8 A0A1A9Y7Z2 A0A087U8W1 A0A1I8Q109 A0A1L8ECH8 A0A1B6CZ84 A0A1I8N0K6 A0A1L8E986 A0A2R5LDU3 A0A293M8Z8 E9H068 A0A2L2YEI8 A0A0P5PVI4 A0A0P6JLX5 A0A1J1IBL5 A0A232F1C2 A0A131YWF7 A0A162NRU7 A0A2R7VTT7 A0A0P4VXY1 A0A0P5YC68 A0A1A9WW31 A0A1E1WW09 A0A131XDA2 A0A1B6MFV0 A0A023G5R3 A0A224YUJ3
A0A182GMI0 A0A1Q3FGE1 J3JXA2 B0WKL7 B0XLX7 A0A0K8URV5 A0A0L7L739 Q7QFV4 A0A182UCW5 A0A182VM34 A0A182LAP0 A0A182WV37 A0A182I8R1 A0A0A1XFM0 Q16XR7 U5EV21 A0A1Y1NCV0 A0A182P765 A0A084VC13 A0A182VXT2 A0A182NFV5 A0A182XXC0 A0A182JNQ3 B4KMD7 B4LNX6 A0A182Q9R6 A0A224XLX3 A0A182RQP5 A0A182JFZ4 W5JHC1 A0A2M4AY75 B4MRP9 A0A1D2NGN4 A0A2P8YKV0 A0A2M4C1G6 A0A069DPU6 A0A0V0G5Q0 A0A0A9W973 A0A1L8DWL4 B3MCL0 D6WK37 A0A1B0G3M5 Q292R7 B4GCX2 A0A0K8TSG8 A0A1L8DWU0 A0A1L8DWP0 A0A0P4VML8 B4J634 A0A1A9UDK1 A0A1W4UME1 A0A1B0GLC6 A0A3B0IYT7 A0A067R8Z8 B4I8U8 A0A0M4EIJ7 A0A3B0IYJ6 Q9W1H9 B3NPU0 A0A0L0CQQ2 A0A2M3ZDG3 B4QB10 B4PA84 A0A336MSR5 A0A1B0D502 A0A1B0BCM8 A0A1A9Y7Z2 A0A087U8W1 A0A1I8Q109 A0A1L8ECH8 A0A1B6CZ84 A0A1I8N0K6 A0A1L8E986 A0A2R5LDU3 A0A293M8Z8 E9H068 A0A2L2YEI8 A0A0P5PVI4 A0A0P6JLX5 A0A1J1IBL5 A0A232F1C2 A0A131YWF7 A0A162NRU7 A0A2R7VTT7 A0A0P4VXY1 A0A0P5YC68 A0A1A9WW31 A0A1E1WW09 A0A131XDA2 A0A1B6MFV0 A0A023G5R3 A0A224YUJ3
Ontologies
GO
Topology
Subcellular location
Nucleus
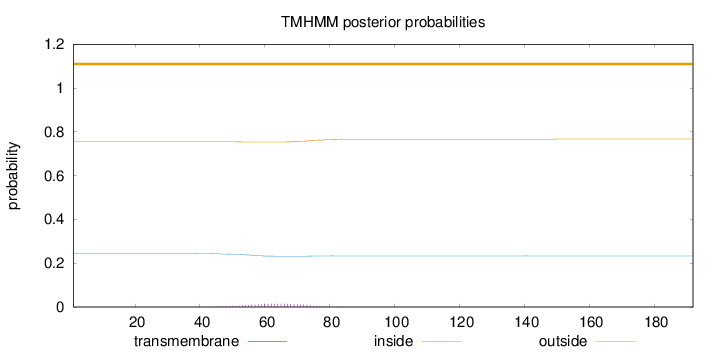
Length:
192
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.32489
Exp number, first 60 AAs:
0.11497
Total prob of N-in:
0.24519
outside
1 - 192
Population Genetic Test Statistics
Pi
130.59664
Theta
136.830433
Tajima's D
0.316169
CLR
0.495138
CSRT
0.457727113644318
Interpretation
Uncertain
