Pre Gene Modal
BGIBMGA011375
Annotation
PREDICTED:_sorting_and_assembly_machinery_component_50_homolog_[Amyelois_transitella]
Full name
Anoctamin
+ More
Sorting and assembly machinery component 50 homolog A
Sorting and assembly machinery component 50 homolog A
Location in the cell
Cytoplasmic Reliability : 1.729 Mitochondrial Reliability : 1.137
Sequence
CDS
ATGGGCACTGTCCACGCGAAGGCAAACCAGCACGATATTGGCGACTCTGGCGGTGGTCTGTTTGTTTTTGAAGAGCCAGTCGAAATGGGCGAGGAACCAATACCAAAACCCACCATCAAATTAGATGGAGTAAGAGCTCGTGTTGATAGAGTGCACGTTGATGGTTTGAATCGTACAAAAGATGACATTATAAAGAATACTATCGACGATCTATTTCACGCAACTGATTTTGAGGATGTTATTGTTAGAGCTCACAAGGTTCGTCGCGCCCTCGATTCAATGGGTTGTTTTCGTGACATCGGTGTGTTCATAGATGTATCGTCCGGACCTGGATCCACACCTGAAGGACTGGAGGTGACGTTTCAAGTTAAAGAGCTATCTCGCGTCATCGGTGGAATAAACACAACAGTCAGCGAGAACGAGGGAAATTTGGTTTTAGGTGTAAAAATGCCAAACATCGCCGGCCGCGGGGAGAAGCTTCAAGCGGAATACAGCATGGGCTATCGAAGCACGTCGAATTTCAATGTCACTGCTACCAAACCGTATCCTCATAAACCCCTCGTGCCTATTGTATCAGCGTCAGTTTATCAGCAGACCCACGAATATCCCTGGTCGGGGTACCGGCTGCTCGACCGGGGTCTGCTAATGGATTTGGCCTTCAAGAGTTCGCCAACGACAAAGCACAACCTTCAGTGGGAGGGCCTGGTGCGCGAGACGGCGGCTCTCAGCAAGACGACCAGCTTCAAAGTCCGCGAGAGCAGCGGTCCTCAGATGAAGTCGATATTGCGACACATCGTGACGGTGGACCACCGGGACGACGCGATCTTCCCGAGTCAGGGCTCCTGGGTGCAGTTCAGCAGCGAGCTGGCCGGGTTGGGCGGGGGCGTGGCCCACCTCAAGACCGAGCTCCAAGCGCAGCAGAACTACGAATTGATCGAGGACATAGTATTGCAGCTCGCCGGAGCCGCAGGGGTTCTGCACGACATGTTCGGGACGGAGATACCTGATCATTTCTTCCTCGGTGGACCCACGACGCTCAGAGGTTTCCAGCAGCGAGGGGTGGGGCCGCACTCGGACGGGCAGGCCACTGGTGGAAAGGTATACTGGGCGGCGGGTGCGCACGTGTACGCGCCGCTGCCCTTCCTTCGGAACAAAAACGGGATTTCGTCCCTGTTCCGGTCGCATTTCTTCGTGAACGCCGGCTGTCTGGCGGCGCCCGATAATGGTTTAACGAATTGGGAATGGGCGAATTCCGCGCGCGTGTCGTGCGGCGCCGGCGTCGCGCTGCGGCTGGGCAGCGTGGCGCGGCTCGAGCTGAACTACTGCGCGCCGCTGAAGGCCCGGACCAACGACTGCACCGCGCACGGCCTCCAGTTCGGGCTCGGAGTGCACTTCTTGTAG
Protein
MGTVHAKANQHDIGDSGGGLFVFEEPVEMGEEPIPKPTIKLDGVRARVDRVHVDGLNRTKDDIIKNTIDDLFHATDFEDVIVRAHKVRRALDSMGCFRDIGVFIDVSSGPGSTPEGLEVTFQVKELSRVIGGINTTVSENEGNLVLGVKMPNIAGRGEKLQAEYSMGYRSTSNFNVTATKPYPHKPLVPIVSASVYQQTHEYPWSGYRLLDRGLLMDLAFKSSPTTKHNLQWEGLVRETAALSKTTSFKVRESSGPQMKSILRHIVTVDHRDDAIFPSQGSWVQFSSELAGLGGGVAHLKTELQAQQNYELIEDIVLQLAGAAGVLHDMFGTEIPDHFFLGGPTTLRGFQQRGVGPHSDGQATGGKVYWAAGAHVYAPLPFLRNKNGISSLFRSHFFVNAGCLAAPDNGLTNWEWANSARVSCGAGVALRLGSVARLELNYCAPLKARTNDCTAHGLQFGLGVHFL
Summary
Description
May play a role in the maintenance of the structure of mitochondrial cristae.
Subunit
Associates with the mitochondrial contact site and cristae organizing system (MICOS) complex (also known as MINOS or MitOS complex).
Similarity
Belongs to the anoctamin family.
Belongs to the SAM50/omp85 family.
Belongs to the SAM50/omp85 family.
Keywords
Complete proteome
Membrane
Mitochondrion
Mitochondrion outer membrane
Reference proteome
Transmembrane
Transmembrane beta strand
Feature
chain Sorting and assembly machinery component 50 homolog A
Uniprot
H9JPC1
A0A2A4JH01
A0A2W1BG45
A0A2A4JFU3
A0A2A4JG65
A0A194PTL7
+ More
A0A194RIN6 A0A2A4JHT8 A0A2W1BMK0 S4PFN9 A0A2H1WPS3 A0A2H1W1M4 A0A067R6V8 A0A2J7RT80 A0A2J7RT84 V5G5V9 A0A139WGS8 A0A1Y1JXG6 J3JTH9 A0A0J7KSU3 A0A232F7K0 K7ILY0 E2A0E1 A0A195FKS2 A0A158NR83 F4WRG8 A0A151IU76 A0A151X596 A0A023GMF1 G3MM13 A0A0K8RIH3 E0W2M2 A0A023FK77 A0A026W5X0 E9IRX5 A0A293MYP5 A0A1E1XFI7 A0A224Z1T2 A0A2P8Y8G2 A0A195BE91 A0A131Z139 A0A0C9SCG9 A0A162CG23 L7M7F0 A0A0P6JZQ7 A0A0P6AP81 A0A131XJR2 A0A088APR0 E9H749 A0A0C9S157 T1JDX0 A0A151IFT7 A0A1B6D716 A0A0L7QQC0 U4UQU1 N6TZY4 A0A1V9XLP2 A0A0M9A1H0 R4WD89 A0A0P5Y9P3 A0A2A3E3P4 A0A1B6M1C4 A0A1D2NFT6 A0A087T813 R4G8S9 A0A0P4VMT8 A0A0V0G3B7 Q4TBF8 A0A3Q1BMH1 A0A224XJ05 F1QBE4 A0A1B6K4W9 A0A087TPN9 A0A2P6KR38 B2GSP6 Q803G5 A0A3B4V834 A0A3B3TBR3 A0A3P8UCP9 A0A1B6HH09 A0A3B4XH90 A0A1A8G6Y9 A0A1A8J5X2 T1HP96 A0A3Q3NPT1 A0A1A7Z6B5 A0A3P9PST6 A0A2U9B273 A0A1A8SH49 A0A1A8N1A4 R7T871 A0A1A8EFJ2 A0A1A8V5V7 A0A3Q3BPR7 A0A3P8PUZ3 A0A146WHB0 A0A3P9D615 A0A3Q3NF96 M4ADD2 A0A3B5KTH1
A0A194RIN6 A0A2A4JHT8 A0A2W1BMK0 S4PFN9 A0A2H1WPS3 A0A2H1W1M4 A0A067R6V8 A0A2J7RT80 A0A2J7RT84 V5G5V9 A0A139WGS8 A0A1Y1JXG6 J3JTH9 A0A0J7KSU3 A0A232F7K0 K7ILY0 E2A0E1 A0A195FKS2 A0A158NR83 F4WRG8 A0A151IU76 A0A151X596 A0A023GMF1 G3MM13 A0A0K8RIH3 E0W2M2 A0A023FK77 A0A026W5X0 E9IRX5 A0A293MYP5 A0A1E1XFI7 A0A224Z1T2 A0A2P8Y8G2 A0A195BE91 A0A131Z139 A0A0C9SCG9 A0A162CG23 L7M7F0 A0A0P6JZQ7 A0A0P6AP81 A0A131XJR2 A0A088APR0 E9H749 A0A0C9S157 T1JDX0 A0A151IFT7 A0A1B6D716 A0A0L7QQC0 U4UQU1 N6TZY4 A0A1V9XLP2 A0A0M9A1H0 R4WD89 A0A0P5Y9P3 A0A2A3E3P4 A0A1B6M1C4 A0A1D2NFT6 A0A087T813 R4G8S9 A0A0P4VMT8 A0A0V0G3B7 Q4TBF8 A0A3Q1BMH1 A0A224XJ05 F1QBE4 A0A1B6K4W9 A0A087TPN9 A0A2P6KR38 B2GSP6 Q803G5 A0A3B4V834 A0A3B3TBR3 A0A3P8UCP9 A0A1B6HH09 A0A3B4XH90 A0A1A8G6Y9 A0A1A8J5X2 T1HP96 A0A3Q3NPT1 A0A1A7Z6B5 A0A3P9PST6 A0A2U9B273 A0A1A8SH49 A0A1A8N1A4 R7T871 A0A1A8EFJ2 A0A1A8V5V7 A0A3Q3BPR7 A0A3P8PUZ3 A0A146WHB0 A0A3P9D615 A0A3Q3NF96 M4ADD2 A0A3B5KTH1
Pubmed
19121390
28756777
26354079
23622113
24845553
18362917
+ More
19820115 28004739 22516182 28648823 20075255 20798317 21347285 21719571 22216098 20566863 24508170 30249741 21282665 28503490 28797301 29403074 26830274 26131772 25576852 28049606 21292972 23537049 28327890 23691247 27289101 27129103 15496914 23594743 29240929 23254933 25186727 23542700
19820115 28004739 22516182 28648823 20075255 20798317 21347285 21719571 22216098 20566863 24508170 30249741 21282665 28503490 28797301 29403074 26830274 26131772 25576852 28049606 21292972 23537049 28327890 23691247 27289101 27129103 15496914 23594743 29240929 23254933 25186727 23542700
EMBL
BABH01009784
BABH01009785
NWSH01001610
PCG70700.1
KZ150513
PZC70633.1
+ More
PCG70699.1 PCG70698.1 KQ459593 KPI96318.1 KQ460152 KPJ17422.1 NWSH01001410 PCG71336.1 KZ149962 PZC76302.1 GAIX01003942 JAA88618.1 ODYU01009771 SOQ54424.1 ODYU01005779 SOQ46995.1 KK852658 KDR19152.1 NEVH01000006 PNF44042.1 PNF44041.1 GALX01003031 JAB65435.1 KQ971345 KYB27071.1 GEZM01102922 JAV51716.1 BT126536 AEE61500.1 LBMM01003554 KMQ93391.1 NNAY01000803 OXU26423.1 GL435586 EFN73078.1 KQ981522 KYN40594.1 ADTU01023820 GL888285 EGI63239.1 KQ980970 KYN10921.1 KQ982540 KYQ55418.1 GBBM01001075 JAC34343.1 JO842914 AEO34531.1 GADI01002851 JAA70957.1 DS235879 EEB19878.1 GBBK01002465 JAC22017.1 KK107390 QOIP01000007 EZA51403.1 RLU20761.1 GL765283 EFZ16741.1 GFWV01019722 MAA44450.1 GFAC01001230 JAT97958.1 GFPF01011979 MAA23125.1 PYGN01000806 PSN40542.1 KQ976504 KYM82888.1 GEDV01004087 JAP84470.1 GBZX01001338 JAG91402.1 LRGB01000872 KZS15432.1 GACK01004813 JAA60221.1 GDIQ01001639 JAN93098.1 GDIP01026602 JAM77113.1 GEFH01001989 JAP66592.1 GL732599 EFX72508.1 GBYB01014461 JAG84228.1 JH432114 KQ977771 KYN00014.1 GEDC01015943 JAS21355.1 KQ414786 KOC60825.1 KB632429 ERL95477.1 APGK01054639 KB741251 ENN71832.1 MNPL01008028 OQR74409.1 KQ435789 KOX74466.1 AK417324 BAN20539.1 GDIP01061060 JAM42655.1 KZ288391 PBC26373.1 GEBQ01010269 JAT29708.1 LJIJ01000053 ODN04148.1 KK113871 KFM61252.1 GAHY01000442 JAA77068.1 GDKW01002808 JAI53787.1 GECL01003512 JAP02612.1 CAAE01007131 CAF89774.1 GFTR01006628 JAW09798.1 BX897714 GECU01037730 GECU01028691 GECU01019340 GECU01001206 GECU01000376 JAS69976.1 JAS79015.1 JAS88366.1 JAT06501.1 JAT07331.1 KK116211 KFM67078.1 MWRG01006701 PRD28768.1 BC165595 AAI65595.1 BC044493 GECU01033760 JAS73946.1 HAEB01020445 HAEC01001405 SBQ66972.1 HAED01018599 HAEE01016906 SBR05044.1 ACPB03012979 ACPB03012980 HADW01013852 HADX01015659 SBP37891.1 CP026244 AWO97878.1 HAEH01012589 HAEI01015083 SBS17552.1 HAEF01021390 HAEG01007168 SBR62549.1 AMQN01003187 AMQN01003188 KB311138 ELT89869.1 HADZ01009803 HAEA01015369 SBQ43849.1 HADY01016843 HAEJ01015637 SBS56094.1 GCES01056922 GCES01056921 JAR29401.1
PCG70699.1 PCG70698.1 KQ459593 KPI96318.1 KQ460152 KPJ17422.1 NWSH01001410 PCG71336.1 KZ149962 PZC76302.1 GAIX01003942 JAA88618.1 ODYU01009771 SOQ54424.1 ODYU01005779 SOQ46995.1 KK852658 KDR19152.1 NEVH01000006 PNF44042.1 PNF44041.1 GALX01003031 JAB65435.1 KQ971345 KYB27071.1 GEZM01102922 JAV51716.1 BT126536 AEE61500.1 LBMM01003554 KMQ93391.1 NNAY01000803 OXU26423.1 GL435586 EFN73078.1 KQ981522 KYN40594.1 ADTU01023820 GL888285 EGI63239.1 KQ980970 KYN10921.1 KQ982540 KYQ55418.1 GBBM01001075 JAC34343.1 JO842914 AEO34531.1 GADI01002851 JAA70957.1 DS235879 EEB19878.1 GBBK01002465 JAC22017.1 KK107390 QOIP01000007 EZA51403.1 RLU20761.1 GL765283 EFZ16741.1 GFWV01019722 MAA44450.1 GFAC01001230 JAT97958.1 GFPF01011979 MAA23125.1 PYGN01000806 PSN40542.1 KQ976504 KYM82888.1 GEDV01004087 JAP84470.1 GBZX01001338 JAG91402.1 LRGB01000872 KZS15432.1 GACK01004813 JAA60221.1 GDIQ01001639 JAN93098.1 GDIP01026602 JAM77113.1 GEFH01001989 JAP66592.1 GL732599 EFX72508.1 GBYB01014461 JAG84228.1 JH432114 KQ977771 KYN00014.1 GEDC01015943 JAS21355.1 KQ414786 KOC60825.1 KB632429 ERL95477.1 APGK01054639 KB741251 ENN71832.1 MNPL01008028 OQR74409.1 KQ435789 KOX74466.1 AK417324 BAN20539.1 GDIP01061060 JAM42655.1 KZ288391 PBC26373.1 GEBQ01010269 JAT29708.1 LJIJ01000053 ODN04148.1 KK113871 KFM61252.1 GAHY01000442 JAA77068.1 GDKW01002808 JAI53787.1 GECL01003512 JAP02612.1 CAAE01007131 CAF89774.1 GFTR01006628 JAW09798.1 BX897714 GECU01037730 GECU01028691 GECU01019340 GECU01001206 GECU01000376 JAS69976.1 JAS79015.1 JAS88366.1 JAT06501.1 JAT07331.1 KK116211 KFM67078.1 MWRG01006701 PRD28768.1 BC165595 AAI65595.1 BC044493 GECU01033760 JAS73946.1 HAEB01020445 HAEC01001405 SBQ66972.1 HAED01018599 HAEE01016906 SBR05044.1 ACPB03012979 ACPB03012980 HADW01013852 HADX01015659 SBP37891.1 CP026244 AWO97878.1 HAEH01012589 HAEI01015083 SBS17552.1 HAEF01021390 HAEG01007168 SBR62549.1 AMQN01003187 AMQN01003188 KB311138 ELT89869.1 HADZ01009803 HAEA01015369 SBQ43849.1 HADY01016843 HAEJ01015637 SBS56094.1 GCES01056922 GCES01056921 JAR29401.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000027135
UP000235965
+ More
UP000007266 UP000036403 UP000215335 UP000002358 UP000000311 UP000078541 UP000005205 UP000007755 UP000078492 UP000075809 UP000009046 UP000053097 UP000279307 UP000245037 UP000078540 UP000076858 UP000005203 UP000000305 UP000078542 UP000053825 UP000030742 UP000019118 UP000192247 UP000053105 UP000242457 UP000094527 UP000054359 UP000007303 UP000257160 UP000000437 UP000261420 UP000261540 UP000265080 UP000261360 UP000015103 UP000261660 UP000242638 UP000246464 UP000014760 UP000264800 UP000265100 UP000265160 UP000261640 UP000002852 UP000261380
UP000007266 UP000036403 UP000215335 UP000002358 UP000000311 UP000078541 UP000005205 UP000007755 UP000078492 UP000075809 UP000009046 UP000053097 UP000279307 UP000245037 UP000078540 UP000076858 UP000005203 UP000000305 UP000078542 UP000053825 UP000030742 UP000019118 UP000192247 UP000053105 UP000242457 UP000094527 UP000054359 UP000007303 UP000257160 UP000000437 UP000261420 UP000261540 UP000265080 UP000261360 UP000015103 UP000261660 UP000242638 UP000246464 UP000014760 UP000264800 UP000265100 UP000265160 UP000261640 UP000002852 UP000261380
Pfam
Interpro
ProteinModelPortal
H9JPC1
A0A2A4JH01
A0A2W1BG45
A0A2A4JFU3
A0A2A4JG65
A0A194PTL7
+ More
A0A194RIN6 A0A2A4JHT8 A0A2W1BMK0 S4PFN9 A0A2H1WPS3 A0A2H1W1M4 A0A067R6V8 A0A2J7RT80 A0A2J7RT84 V5G5V9 A0A139WGS8 A0A1Y1JXG6 J3JTH9 A0A0J7KSU3 A0A232F7K0 K7ILY0 E2A0E1 A0A195FKS2 A0A158NR83 F4WRG8 A0A151IU76 A0A151X596 A0A023GMF1 G3MM13 A0A0K8RIH3 E0W2M2 A0A023FK77 A0A026W5X0 E9IRX5 A0A293MYP5 A0A1E1XFI7 A0A224Z1T2 A0A2P8Y8G2 A0A195BE91 A0A131Z139 A0A0C9SCG9 A0A162CG23 L7M7F0 A0A0P6JZQ7 A0A0P6AP81 A0A131XJR2 A0A088APR0 E9H749 A0A0C9S157 T1JDX0 A0A151IFT7 A0A1B6D716 A0A0L7QQC0 U4UQU1 N6TZY4 A0A1V9XLP2 A0A0M9A1H0 R4WD89 A0A0P5Y9P3 A0A2A3E3P4 A0A1B6M1C4 A0A1D2NFT6 A0A087T813 R4G8S9 A0A0P4VMT8 A0A0V0G3B7 Q4TBF8 A0A3Q1BMH1 A0A224XJ05 F1QBE4 A0A1B6K4W9 A0A087TPN9 A0A2P6KR38 B2GSP6 Q803G5 A0A3B4V834 A0A3B3TBR3 A0A3P8UCP9 A0A1B6HH09 A0A3B4XH90 A0A1A8G6Y9 A0A1A8J5X2 T1HP96 A0A3Q3NPT1 A0A1A7Z6B5 A0A3P9PST6 A0A2U9B273 A0A1A8SH49 A0A1A8N1A4 R7T871 A0A1A8EFJ2 A0A1A8V5V7 A0A3Q3BPR7 A0A3P8PUZ3 A0A146WHB0 A0A3P9D615 A0A3Q3NF96 M4ADD2 A0A3B5KTH1
A0A194RIN6 A0A2A4JHT8 A0A2W1BMK0 S4PFN9 A0A2H1WPS3 A0A2H1W1M4 A0A067R6V8 A0A2J7RT80 A0A2J7RT84 V5G5V9 A0A139WGS8 A0A1Y1JXG6 J3JTH9 A0A0J7KSU3 A0A232F7K0 K7ILY0 E2A0E1 A0A195FKS2 A0A158NR83 F4WRG8 A0A151IU76 A0A151X596 A0A023GMF1 G3MM13 A0A0K8RIH3 E0W2M2 A0A023FK77 A0A026W5X0 E9IRX5 A0A293MYP5 A0A1E1XFI7 A0A224Z1T2 A0A2P8Y8G2 A0A195BE91 A0A131Z139 A0A0C9SCG9 A0A162CG23 L7M7F0 A0A0P6JZQ7 A0A0P6AP81 A0A131XJR2 A0A088APR0 E9H749 A0A0C9S157 T1JDX0 A0A151IFT7 A0A1B6D716 A0A0L7QQC0 U4UQU1 N6TZY4 A0A1V9XLP2 A0A0M9A1H0 R4WD89 A0A0P5Y9P3 A0A2A3E3P4 A0A1B6M1C4 A0A1D2NFT6 A0A087T813 R4G8S9 A0A0P4VMT8 A0A0V0G3B7 Q4TBF8 A0A3Q1BMH1 A0A224XJ05 F1QBE4 A0A1B6K4W9 A0A087TPN9 A0A2P6KR38 B2GSP6 Q803G5 A0A3B4V834 A0A3B3TBR3 A0A3P8UCP9 A0A1B6HH09 A0A3B4XH90 A0A1A8G6Y9 A0A1A8J5X2 T1HP96 A0A3Q3NPT1 A0A1A7Z6B5 A0A3P9PST6 A0A2U9B273 A0A1A8SH49 A0A1A8N1A4 R7T871 A0A1A8EFJ2 A0A1A8V5V7 A0A3Q3BPR7 A0A3P8PUZ3 A0A146WHB0 A0A3P9D615 A0A3Q3NF96 M4ADD2 A0A3B5KTH1
PDB
5D0Q
E-value=2.68134e-08,
Score=140
Ontologies
GO
Topology
Subcellular location
Membrane
Mitochondrion outer membrane
Mitochondrion outer membrane
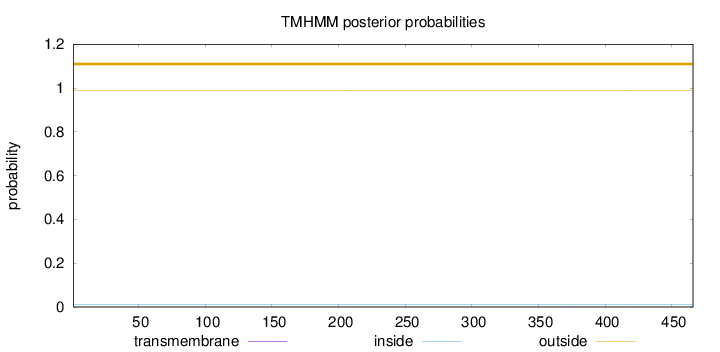
Length:
466
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00652
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01056
outside
1 - 466
Population Genetic Test Statistics
Pi
209.112193
Theta
145.08063
Tajima's D
1.471363
CLR
1.172408
CSRT
0.781560921953902
Interpretation
Uncertain
