Pre Gene Modal
BGIBMGA011230
Annotation
PREDICTED:_putative_tRNA_pseudouridine_synthase_Pus10_[Papilio_polytes]
Full name
Putative tRNA pseudouridine synthase Pus10
Alternative Name
Coiled-coil domain-containing protein 139 homolog
tRNA pseudouridine 55 synthase
tRNA pseudouridylate synthase
tRNA-uridine isomerase
tRNA pseudouridine 55 synthase
tRNA pseudouridylate synthase
tRNA-uridine isomerase
Location in the cell
Nuclear Reliability : 3.034
Sequence
CDS
ATGGACAATAAAGCCATCGTTAAATTCTGTAAAGAGGTAGGGTGTTGCGATATATGCTGCCTGCGTTATTTAGGGCTTAAGAACCCGGCAACTTACGAAAACTACAAAGAATACATACATAAATATACAGAAAGCGGTGTAGAAGTAGAAAATAATTCAGAAGAACAACAAATCACTAGCAATCCAGTAGACAATATAAATGGCAATAAAGATGTCAATGGCTGTGATGATGAACCACCGAAAAAGAAAAAGAAGATATCGATATGCGTAAGCTGTTTAGGTATACTCCAGGAAGAGAACTGGTCTGAATGCTTTAATATGGTCAAAGAGACTTTGGAGAAAAAACGTTACGAATGCTCAACTTTTGCATGTGCACTCTCTGCTCCGATCGCTACATTATTACGAGATAAAGCCATAATCCTACACCTGAGTGATGCCTTTAAAGATTATAAAGAAGATACCTTAACACCACTGAAGGAAGCATGGAAATGGACGTTTGGTGTCAAACTTGCCGAACAAATAGGTATGACACTAGATTCCGGAGCCATTTCACCGTTACTGATTACACTTAATATGGAGTATCCTGATGAACCACAGGAACTAGAAATCTTAAAGAAATTATCACCATCATTGTTCCAAGAGAGACAGAAGCAGAGAAGGAGGTTCACGGTTGAGTTTACCCGGCCATCAGTAGAACAGGCCATGGACAAGATTAGTCTTGAGCAGTTGAAGGGTGTCGATAAGTGGAATAAAATTCCATCGGTGGACATTGAAGCTAAGTGTGTCAGTGCTATGTGCGTGCATGCCCCTTCTTATCTGGGGGGTAGGTATATAAAATTGAGCAGAGAGCTTCCGCAGACGCCCTGGCTGATCAAGGGCAAGAGGATGATGGAATCATCCGTCCAGGAGATCTTATTCGAACCCATAGCAAAAGTATTTGACTTGACACCTGAAGACGTGGACCACAGATTGAAGTTCATGTCGGCTGGACGCGAAGACGTGGACGTGAGGTGTCTCGGTGATGGTAGACCCTTCGCTATCGAAATCACAGACCCGAGAAGAGACCTCACCGGTGACGAGCTGAAGAGGGTCTGCAAGGAGATTTCGAAAGGTGGCCAAGTGATCGTTCAGAAACTAATGCATGTCTCCAGGAACGAGTTGTCGGAACTTAAAAAGGGGGAAGAGACGAAATGCAAGACTTACGAAGCTCTGTGCATAAAGCTCGGAAATGAGAACGCTGAACCGAATATTCTGAATGGAAAAACGCCTGTCGCCGTCACCGAGGAGGACATCCAGAGGATAAACGAGTACAGGAACACCGAGGCAGGGGACGAGGCCAGGATCGTGGTGAAACAGAGAACCCCGATCCGAGTGCTGCACAGGCGCCCGCTGCTCACCAGGACCCGGAGGATATATGACGTAGTGGCCAGGAGGGTGCCTGACTACCCGCAATTGTTCACCCTCACAATCCGCACGGAAGCGGGCACGTACGTGAAGGAGTGGTGCCACGGGGAGTTCGGTCGCACCACGCCGAGCCTCCGCGACGCCGCGGGCTTCCGGGCCGACATACTGGCGCTCGACGTCACCGCGGTGCTGCTCGACTGGCCGCGACCCCAATAA
Protein
MDNKAIVKFCKEVGCCDICCLRYLGLKNPATYENYKEYIHKYTESGVEVENNSEEQQITSNPVDNINGNKDVNGCDDEPPKKKKKISICVSCLGILQEENWSECFNMVKETLEKKRYECSTFACALSAPIATLLRDKAIILHLSDAFKDYKEDTLTPLKEAWKWTFGVKLAEQIGMTLDSGAISPLLITLNMEYPDEPQELEILKKLSPSLFQERQKQRRRFTVEFTRPSVEQAMDKISLEQLKGVDKWNKIPSVDIEAKCVSAMCVHAPSYLGGRYIKLSRELPQTPWLIKGKRMMESSVQEILFEPIAKVFDLTPEDVDHRLKFMSAGREDVDVRCLGDGRPFAIEITDPRRDLTGDELKRVCKEISKGGQVIVQKLMHVSRNELSELKKGEETKCKTYEALCIKLGNENAEPNILNGKTPVAVTEEDIQRINEYRNTEAGDEARIVVKQRTPIRVLHRRPLLTRTRRIYDVVARRVPDYPQLFTLTIRTEAGTYVKEWCHGEFGRTTPSLRDAAGFRADILALDVTAVLLDWPRPQ
Summary
Description
Responsible for synthesis of pseudouridine from uracil-55 in the psi GC loop of transfer RNAs.
Catalytic Activity
uridine(55) in tRNA = pseudouridine(55) in tRNA
Similarity
Belongs to the pseudouridine synthase Pus10 family.
Keywords
Coiled coil
Complete proteome
Isomerase
Reference proteome
tRNA processing
Feature
chain Putative tRNA pseudouridine synthase Pus10
Uniprot
H9JNX6
A0A2H1W737
A0A194PU88
A0A2W1B7M0
A0A194RN45
A0A212EVT9
+ More
A0A212FD27 A0A1Y1NEJ9 A0A0P4VQJ0 E0VC88 T1IET3 A0A2P8YXZ2 A0A0V0GCK4 A0A2J7R5R5 A0A023F1C6 A0A0P5UN82 A0A0P5VMG4 A0A0P5THR2 A0A0N8CQF7 A0A0P5QCS6 A0A0N8CJP9 A0A0P6BZS9 A0A0N8BVC1 A0A067QIG3 A0A088A116 A0A0P4XL53 A0A0P5D9R3 A0A0P5T5T9 A0A2C9LBZ3 A0A0M9A7V8 A0A0N8CHR3 A0A2A3ENK5 A0A131YTX1 A0A210QXC8 A0A0K8SL69 A0A0A9XA35 A0A0K2SW20 A0A224Z8T0 A0A0L7QUG7 A0A0N8BYD3 A0A1B6E3L2 Q16T84 A0A3L8DGS4 A0A1Y1Y3W1 A0A182HD50 A0A2R5LC42 B4Q5N1 B4ICS7 A0A1B6HWQ8 A0A182GY00 A0A158NQG7 A0A195ET53 F4X4U5 G3MHB3 T1J968 Q9VPK7 A0A084W4F3 A0A151HYG8 A0A151X633 A0A1U7UW97 A0A3B3RTB8 A0A182LXM9 A0A336MFF3 A0A2T7NC28 A0A1W4V331 A0A182RMR3 A0A1E1XG78 B3MUD0 D6W8D0 A0A182IQ80 A0A0L8HXZ6 A0A0A1WRW9 A0A091CUA5 G5C1Z8 A0A226EQW2 G1REG8 M3ZC52 A0A2J8XCI8 E2R061 A0A3Q7RRM1 A0A3Q2SQW1 A0A034V6E1 A0A182Q9H7 A0A0D9RPN6 A0A250YJP2 G7PM98 M3W8T7 A0A147A231 A0A182XFR1 A0A2R8ZVY3 A0A151IC49 A0A096NQX3 A0A2K5P869 F6U5F7 A0A2I3S2F4 A0A3B3BLK7 A0A2I3LIC8 A0A2K5P8D6 A0A2R8ZQ34
A0A212FD27 A0A1Y1NEJ9 A0A0P4VQJ0 E0VC88 T1IET3 A0A2P8YXZ2 A0A0V0GCK4 A0A2J7R5R5 A0A023F1C6 A0A0P5UN82 A0A0P5VMG4 A0A0P5THR2 A0A0N8CQF7 A0A0P5QCS6 A0A0N8CJP9 A0A0P6BZS9 A0A0N8BVC1 A0A067QIG3 A0A088A116 A0A0P4XL53 A0A0P5D9R3 A0A0P5T5T9 A0A2C9LBZ3 A0A0M9A7V8 A0A0N8CHR3 A0A2A3ENK5 A0A131YTX1 A0A210QXC8 A0A0K8SL69 A0A0A9XA35 A0A0K2SW20 A0A224Z8T0 A0A0L7QUG7 A0A0N8BYD3 A0A1B6E3L2 Q16T84 A0A3L8DGS4 A0A1Y1Y3W1 A0A182HD50 A0A2R5LC42 B4Q5N1 B4ICS7 A0A1B6HWQ8 A0A182GY00 A0A158NQG7 A0A195ET53 F4X4U5 G3MHB3 T1J968 Q9VPK7 A0A084W4F3 A0A151HYG8 A0A151X633 A0A1U7UW97 A0A3B3RTB8 A0A182LXM9 A0A336MFF3 A0A2T7NC28 A0A1W4V331 A0A182RMR3 A0A1E1XG78 B3MUD0 D6W8D0 A0A182IQ80 A0A0L8HXZ6 A0A0A1WRW9 A0A091CUA5 G5C1Z8 A0A226EQW2 G1REG8 M3ZC52 A0A2J8XCI8 E2R061 A0A3Q7RRM1 A0A3Q2SQW1 A0A034V6E1 A0A182Q9H7 A0A0D9RPN6 A0A250YJP2 G7PM98 M3W8T7 A0A147A231 A0A182XFR1 A0A2R8ZVY3 A0A151IC49 A0A096NQX3 A0A2K5P869 F6U5F7 A0A2I3S2F4 A0A3B3BLK7 A0A2I3LIC8 A0A2K5P8D6 A0A2R8ZQ34
EC Number
5.4.99.25
Pubmed
19121390
26354079
28756777
22118469
28004739
27129103
+ More
20566863 29403074 25474469 24845553 15562597 26830274 28812685 25401762 28797301 17510324 30249741 26483478 17994087 22936249 21347285 21719571 22216098 10731132 12537572 12537569 24438588 29240929 28503490 18362917 19820115 25830018 21993625 16341006 25348373 28087693 22002653 17975172 22722832 17431167 16136131 29451363
20566863 29403074 25474469 24845553 15562597 26830274 28812685 25401762 28797301 17510324 30249741 26483478 17994087 22936249 21347285 21719571 22216098 10731132 12537572 12537569 24438588 29240929 28503490 18362917 19820115 25830018 21993625 16341006 25348373 28087693 22002653 17975172 22722832 17431167 16136131 29451363
EMBL
BABH01009782
BABH01009783
BABH01009784
ODYU01006767
SOQ48915.1
KQ459593
+ More
KPI96319.1 KZ150714 PZC70364.1 KQ460152 KPJ17421.1 AGBW02012126 OWR45599.1 AGBW02009108 OWR51656.1 GEZM01005649 JAV96008.1 GDKW01001317 JAI55278.1 DS235049 EEB10994.1 ACPB03014909 PYGN01000293 PSN49117.1 GECL01001026 JAP05098.1 NEVH01006990 PNF36171.1 GBBI01003896 JAC14816.1 GDIP01110621 JAL93093.1 GDIP01098054 LRGB01000311 JAM05661.1 KZS19858.1 GDIP01131896 JAL71818.1 GDIP01108991 JAL94723.1 GDIQ01116152 GDIP01140510 JAL35574.1 JAL63204.1 GDIP01125055 JAL78659.1 GDIP01008130 JAM95585.1 GDIQ01141291 JAL10435.1 KK853541 KDR06864.1 GDIP01239837 JAI83564.1 GDIP01164499 JAJ58903.1 GDIP01130544 JAL73170.1 KQ435716 KOX79040.1 GDIP01130543 JAL73171.1 KZ288215 PBC32621.1 GEDV01006150 JAP82407.1 NEDP02001369 OWF53363.1 GBRD01011768 JAG54056.1 GBHO01029639 JAG13965.1 HACA01000354 CDW17715.1 GFPF01012875 MAA24021.1 KQ414735 KOC62260.1 GDIQ01132795 JAL18931.1 GEDC01004790 JAS32508.1 CH477658 EAT37670.1 QOIP01000009 RLU19068.1 MCFE01000269 ORX92575.1 JXUM01033969 KQ561007 KXJ80059.1 GGLE01002903 MBY07029.1 CM000361 CM002910 EDX03181.1 KMY87250.1 CH480829 EDW45353.1 GECU01028641 JAS79065.1 JXUM01096545 KQ564272 KXJ72497.1 ADTU01023318 KQ981993 KYN31092.1 GL888679 EGI58518.1 JO841264 AEO32881.1 JH431970 AE014134 AY069446 ATLV01020330 KE525298 KFB45097.1 KQ976730 KYM76432.1 KQ982491 KYQ55760.1 AXCM01001492 UFQS01000836 UFQT01000836 SSX07180.1 SSX27523.1 PZQS01000014 PVD18724.1 GFAC01000925 JAT98263.1 CH902624 EDV33459.2 KQ971307 EFA10881.1 KQ417037 KOF94087.1 GBXI01013032 JAD01260.1 KN124472 KFO21180.1 JH172942 GEBF01006669 EHB15559.1 JAN96963.1 LNIX01000002 OXA59434.1 ADFV01009950 ADFV01009951 ADFV01009952 ADFV01009953 ADFV01009954 ADFV01009955 ABGA01395332 ABGA01395333 ABGA01395334 ABGA01395335 ABGA01395336 ABGA01395337 ABGA01395338 NDHI03003368 PNJ79750.1 PNJ79753.1 AAEX03007551 GAKP01020076 JAC38876.1 AXCN02001803 AQIB01041624 AQIB01041625 GFFW01001029 JAV43759.1 CM001288 EHH55593.1 AANG04002548 GCES01014101 JAR72222.1 AJFE02109096 AJFE02109097 AJFE02109098 AJFE02109099 AJFE02109100 AJFE02109101 KQ978069 KYM97355.1 AHZZ02005464 AHZZ02005465 JSUE03010979 JSUE03010980 JSUE03010981 CM001265 EHH22144.1 AACZ04063513
KPI96319.1 KZ150714 PZC70364.1 KQ460152 KPJ17421.1 AGBW02012126 OWR45599.1 AGBW02009108 OWR51656.1 GEZM01005649 JAV96008.1 GDKW01001317 JAI55278.1 DS235049 EEB10994.1 ACPB03014909 PYGN01000293 PSN49117.1 GECL01001026 JAP05098.1 NEVH01006990 PNF36171.1 GBBI01003896 JAC14816.1 GDIP01110621 JAL93093.1 GDIP01098054 LRGB01000311 JAM05661.1 KZS19858.1 GDIP01131896 JAL71818.1 GDIP01108991 JAL94723.1 GDIQ01116152 GDIP01140510 JAL35574.1 JAL63204.1 GDIP01125055 JAL78659.1 GDIP01008130 JAM95585.1 GDIQ01141291 JAL10435.1 KK853541 KDR06864.1 GDIP01239837 JAI83564.1 GDIP01164499 JAJ58903.1 GDIP01130544 JAL73170.1 KQ435716 KOX79040.1 GDIP01130543 JAL73171.1 KZ288215 PBC32621.1 GEDV01006150 JAP82407.1 NEDP02001369 OWF53363.1 GBRD01011768 JAG54056.1 GBHO01029639 JAG13965.1 HACA01000354 CDW17715.1 GFPF01012875 MAA24021.1 KQ414735 KOC62260.1 GDIQ01132795 JAL18931.1 GEDC01004790 JAS32508.1 CH477658 EAT37670.1 QOIP01000009 RLU19068.1 MCFE01000269 ORX92575.1 JXUM01033969 KQ561007 KXJ80059.1 GGLE01002903 MBY07029.1 CM000361 CM002910 EDX03181.1 KMY87250.1 CH480829 EDW45353.1 GECU01028641 JAS79065.1 JXUM01096545 KQ564272 KXJ72497.1 ADTU01023318 KQ981993 KYN31092.1 GL888679 EGI58518.1 JO841264 AEO32881.1 JH431970 AE014134 AY069446 ATLV01020330 KE525298 KFB45097.1 KQ976730 KYM76432.1 KQ982491 KYQ55760.1 AXCM01001492 UFQS01000836 UFQT01000836 SSX07180.1 SSX27523.1 PZQS01000014 PVD18724.1 GFAC01000925 JAT98263.1 CH902624 EDV33459.2 KQ971307 EFA10881.1 KQ417037 KOF94087.1 GBXI01013032 JAD01260.1 KN124472 KFO21180.1 JH172942 GEBF01006669 EHB15559.1 JAN96963.1 LNIX01000002 OXA59434.1 ADFV01009950 ADFV01009951 ADFV01009952 ADFV01009953 ADFV01009954 ADFV01009955 ABGA01395332 ABGA01395333 ABGA01395334 ABGA01395335 ABGA01395336 ABGA01395337 ABGA01395338 NDHI03003368 PNJ79750.1 PNJ79753.1 AAEX03007551 GAKP01020076 JAC38876.1 AXCN02001803 AQIB01041624 AQIB01041625 GFFW01001029 JAV43759.1 CM001288 EHH55593.1 AANG04002548 GCES01014101 JAR72222.1 AJFE02109096 AJFE02109097 AJFE02109098 AJFE02109099 AJFE02109100 AJFE02109101 KQ978069 KYM97355.1 AHZZ02005464 AHZZ02005465 JSUE03010979 JSUE03010980 JSUE03010981 CM001265 EHH22144.1 AACZ04063513
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000009046
UP000015103
+ More
UP000245037 UP000235965 UP000076858 UP000027135 UP000005203 UP000076420 UP000053105 UP000242457 UP000242188 UP000053825 UP000008820 UP000279307 UP000193498 UP000069940 UP000249989 UP000000304 UP000001292 UP000005205 UP000078541 UP000007755 UP000000803 UP000030765 UP000078540 UP000075809 UP000189704 UP000261540 UP000075883 UP000245119 UP000192221 UP000075900 UP000007801 UP000007266 UP000075880 UP000053454 UP000028990 UP000006813 UP000198287 UP000001073 UP000001595 UP000002254 UP000286640 UP000265000 UP000075886 UP000029965 UP000009130 UP000011712 UP000076407 UP000240080 UP000078542 UP000028761 UP000233060 UP000006718 UP000002277 UP000261560
UP000245037 UP000235965 UP000076858 UP000027135 UP000005203 UP000076420 UP000053105 UP000242457 UP000242188 UP000053825 UP000008820 UP000279307 UP000193498 UP000069940 UP000249989 UP000000304 UP000001292 UP000005205 UP000078541 UP000007755 UP000000803 UP000030765 UP000078540 UP000075809 UP000189704 UP000261540 UP000075883 UP000245119 UP000192221 UP000075900 UP000007801 UP000007266 UP000075880 UP000053454 UP000028990 UP000006813 UP000198287 UP000001073 UP000001595 UP000002254 UP000286640 UP000265000 UP000075886 UP000029965 UP000009130 UP000011712 UP000076407 UP000240080 UP000078542 UP000028761 UP000233060 UP000006718 UP000002277 UP000261560
SUPFAM
SSF55120
SSF55120
ProteinModelPortal
H9JNX6
A0A2H1W737
A0A194PU88
A0A2W1B7M0
A0A194RN45
A0A212EVT9
+ More
A0A212FD27 A0A1Y1NEJ9 A0A0P4VQJ0 E0VC88 T1IET3 A0A2P8YXZ2 A0A0V0GCK4 A0A2J7R5R5 A0A023F1C6 A0A0P5UN82 A0A0P5VMG4 A0A0P5THR2 A0A0N8CQF7 A0A0P5QCS6 A0A0N8CJP9 A0A0P6BZS9 A0A0N8BVC1 A0A067QIG3 A0A088A116 A0A0P4XL53 A0A0P5D9R3 A0A0P5T5T9 A0A2C9LBZ3 A0A0M9A7V8 A0A0N8CHR3 A0A2A3ENK5 A0A131YTX1 A0A210QXC8 A0A0K8SL69 A0A0A9XA35 A0A0K2SW20 A0A224Z8T0 A0A0L7QUG7 A0A0N8BYD3 A0A1B6E3L2 Q16T84 A0A3L8DGS4 A0A1Y1Y3W1 A0A182HD50 A0A2R5LC42 B4Q5N1 B4ICS7 A0A1B6HWQ8 A0A182GY00 A0A158NQG7 A0A195ET53 F4X4U5 G3MHB3 T1J968 Q9VPK7 A0A084W4F3 A0A151HYG8 A0A151X633 A0A1U7UW97 A0A3B3RTB8 A0A182LXM9 A0A336MFF3 A0A2T7NC28 A0A1W4V331 A0A182RMR3 A0A1E1XG78 B3MUD0 D6W8D0 A0A182IQ80 A0A0L8HXZ6 A0A0A1WRW9 A0A091CUA5 G5C1Z8 A0A226EQW2 G1REG8 M3ZC52 A0A2J8XCI8 E2R061 A0A3Q7RRM1 A0A3Q2SQW1 A0A034V6E1 A0A182Q9H7 A0A0D9RPN6 A0A250YJP2 G7PM98 M3W8T7 A0A147A231 A0A182XFR1 A0A2R8ZVY3 A0A151IC49 A0A096NQX3 A0A2K5P869 F6U5F7 A0A2I3S2F4 A0A3B3BLK7 A0A2I3LIC8 A0A2K5P8D6 A0A2R8ZQ34
A0A212FD27 A0A1Y1NEJ9 A0A0P4VQJ0 E0VC88 T1IET3 A0A2P8YXZ2 A0A0V0GCK4 A0A2J7R5R5 A0A023F1C6 A0A0P5UN82 A0A0P5VMG4 A0A0P5THR2 A0A0N8CQF7 A0A0P5QCS6 A0A0N8CJP9 A0A0P6BZS9 A0A0N8BVC1 A0A067QIG3 A0A088A116 A0A0P4XL53 A0A0P5D9R3 A0A0P5T5T9 A0A2C9LBZ3 A0A0M9A7V8 A0A0N8CHR3 A0A2A3ENK5 A0A131YTX1 A0A210QXC8 A0A0K8SL69 A0A0A9XA35 A0A0K2SW20 A0A224Z8T0 A0A0L7QUG7 A0A0N8BYD3 A0A1B6E3L2 Q16T84 A0A3L8DGS4 A0A1Y1Y3W1 A0A182HD50 A0A2R5LC42 B4Q5N1 B4ICS7 A0A1B6HWQ8 A0A182GY00 A0A158NQG7 A0A195ET53 F4X4U5 G3MHB3 T1J968 Q9VPK7 A0A084W4F3 A0A151HYG8 A0A151X633 A0A1U7UW97 A0A3B3RTB8 A0A182LXM9 A0A336MFF3 A0A2T7NC28 A0A1W4V331 A0A182RMR3 A0A1E1XG78 B3MUD0 D6W8D0 A0A182IQ80 A0A0L8HXZ6 A0A0A1WRW9 A0A091CUA5 G5C1Z8 A0A226EQW2 G1REG8 M3ZC52 A0A2J8XCI8 E2R061 A0A3Q7RRM1 A0A3Q2SQW1 A0A034V6E1 A0A182Q9H7 A0A0D9RPN6 A0A250YJP2 G7PM98 M3W8T7 A0A147A231 A0A182XFR1 A0A2R8ZVY3 A0A151IC49 A0A096NQX3 A0A2K5P869 F6U5F7 A0A2I3S2F4 A0A3B3BLK7 A0A2I3LIC8 A0A2K5P8D6 A0A2R8ZQ34
PDB
2V9K
E-value=5.76085e-56,
Score=552
Ontologies
GO
PANTHER
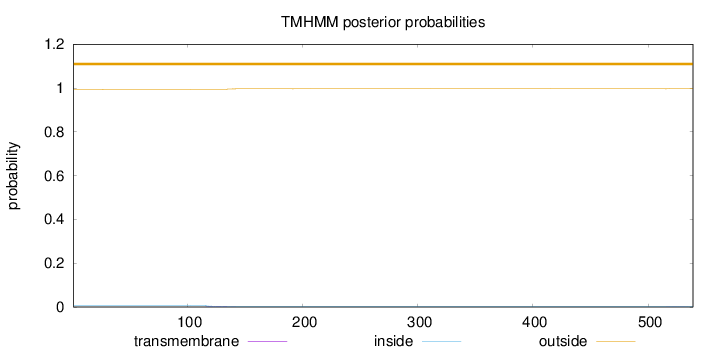
Topology
Length:
539
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0690099999999998
Exp number, first 60 AAs:
0.00137
Total prob of N-in:
0.00703
outside
1 - 539
Population Genetic Test Statistics
Pi
197.31171
Theta
129.247114
Tajima's D
0.857205
CLR
1.323146
CSRT
0.619719014049298
Interpretation
Uncertain
