Gene
KWMTBOMO14054
Annotation
presenilin_enhancer_[Bombyx_mori]
Full name
Gamma-secretase subunit pen-2
Alternative Name
Presenilin enhancer protein 2
Location in the cell
PlasmaMembrane Reliability : 3.833
Sequence
CDS
ATGGATTTAAGTAAACTGCCGAACGATATGAAACTACAATTGTGCAAATCATATTTTAAAGTTGGTTGTCTGTTCCTACCGTTTGTGTGGGCGGTGAATGTGTGTTGGTTCTTCCGAGATGCATTTATAAAACCAGCATATGATGAGCAGAAACTAATAAAGAAATTTGTGATAATGAGTGCAATTGGAGCTTTCGTGTGGGCTGTTATATTGGTTACGTGGATTACGGTGTATCAAACAAACAGAGTGTCGTGGGGAGCTACAGGCGACGCACTATCCTTCATAGTACCTTTAGGACGAGCCTAG
Protein
MDLSKLPNDMKLQLCKSYFKVGCLFLPFVWAVNVCWFFRDAFIKPAYDEQKLIKKFVIMSAIGAFVWAVILVTWITVYQTNRVSWGATGDALSFIVPLGRA
Summary
Description
Essential subunit of the gamma-secretase complex, an endoprotease complex that catalyzes the intramembrane cleavage of integral membrane proteins such as Notch. It probably represents the last step of maturation of gamma-secretase, facilitating endoproteolysis of presenilin and conferring gamma-secretase activity.
Subunit
Component of the gamma-secretase complex, a complex composed of a presenilin (Psn) homodimer, nicastrin (Nct), Aph-1 and Pen-2.
Similarity
Belongs to the PEN-2 family.
Keywords
Complete proteome
Membrane
Notch signaling pathway
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Gamma-secretase subunit pen-2
Uniprot
Q2F5X1
A0A1E1WHM4
A0A194PSA5
A0A2H1VK38
A0A2W1BMJ0
A0A3S2M6R6
+ More
S4P4B3 A0A194RJ04 A0A2A4K5L7 A0A212EYA0 B4LNL7 B3MBC9 A0A1W4UJV8 B4NN19 B4KM71 B4J5R5 B3NMP6 B4NSL5 A0A1Q3G2Q4 Q640D2 B4QCC0 A0A0B4LFM5 B4P4N7 Q86BE9 Q28GF7 A0A0N7Z955 R4G877 Q5PQA0 Q178A8 B4HNL1 U5EVA9 A0A0K8TPH9 E2BQH0 A0A067QRX9 A0A0K8VVP3 A0A023ECS1 A0A0C9QYR3 A0A224Y1G1 A0A0V0GB54 B5DZQ9 B4GIQ3 A0A2G9QEV9 A0A385JF77 A0A3B0JKK7 A0A1X9IXE3 A0A0A9VWP5 A0A088AD44 E0VLA5 A0A2A3ELV5 V9IHQ6 A0A1B6J8L5 R4V2K3 A0A1B6FN65 A0A182MW29 A0A158NK86 A0A139WGF1 A0A182JU72 A0A182QE36 A0A023FD68 A0A182NUB4 A0A2J7RA64 A0A1A9WJE2 W5JCM5 A0A195EHR5 F4W8I4 A0A182SWV9 A0A2M3ZLE9 A0A182Y057 A0A151I7U1 A0A182VK82 A0A182TTD2 A0A182L1J5 Q7Q3Y1 A0A2M4C4C1 A0A182RZT2 A0A182W754 A0A1B0BT68 A0A3B4G386 A0A3Q2VAI7 A0A3Q4HTN4 A0A3P8N743 A0A3P9C1U2 A0A224Z4M2 A0A131YIU7 L7M5X3 A0A131XHN3 A0A182PMN5 A0A182IKZ2 A0A1A9YG60 B6DDY1 A0A3Q3MLH6 A0A2M4AWT9 A0A182F3E4 A0A1I8N311 A0A1L8D8T5 A0A1B6M571 A0A023GDT2 G3MN57 A0A026X2M0 A0A3Q1HYZ0
S4P4B3 A0A194RJ04 A0A2A4K5L7 A0A212EYA0 B4LNL7 B3MBC9 A0A1W4UJV8 B4NN19 B4KM71 B4J5R5 B3NMP6 B4NSL5 A0A1Q3G2Q4 Q640D2 B4QCC0 A0A0B4LFM5 B4P4N7 Q86BE9 Q28GF7 A0A0N7Z955 R4G877 Q5PQA0 Q178A8 B4HNL1 U5EVA9 A0A0K8TPH9 E2BQH0 A0A067QRX9 A0A0K8VVP3 A0A023ECS1 A0A0C9QYR3 A0A224Y1G1 A0A0V0GB54 B5DZQ9 B4GIQ3 A0A2G9QEV9 A0A385JF77 A0A3B0JKK7 A0A1X9IXE3 A0A0A9VWP5 A0A088AD44 E0VLA5 A0A2A3ELV5 V9IHQ6 A0A1B6J8L5 R4V2K3 A0A1B6FN65 A0A182MW29 A0A158NK86 A0A139WGF1 A0A182JU72 A0A182QE36 A0A023FD68 A0A182NUB4 A0A2J7RA64 A0A1A9WJE2 W5JCM5 A0A195EHR5 F4W8I4 A0A182SWV9 A0A2M3ZLE9 A0A182Y057 A0A151I7U1 A0A182VK82 A0A182TTD2 A0A182L1J5 Q7Q3Y1 A0A2M4C4C1 A0A182RZT2 A0A182W754 A0A1B0BT68 A0A3B4G386 A0A3Q2VAI7 A0A3Q4HTN4 A0A3P8N743 A0A3P9C1U2 A0A224Z4M2 A0A131YIU7 L7M5X3 A0A131XHN3 A0A182PMN5 A0A182IKZ2 A0A1A9YG60 B6DDY1 A0A3Q3MLH6 A0A2M4AWT9 A0A182F3E4 A0A1I8N311 A0A1L8D8T5 A0A1B6M571 A0A023GDT2 G3MN57 A0A026X2M0 A0A3Q1HYZ0
Pubmed
26354079
28756777
23622113
22118469
17994087
22936249
+ More
27762356 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 12110170 12660785 20431018 27129103 17510324 26369729 20798317 24845553 24945155 15632085 27966611 25401762 26823975 20566863 21347285 18362917 19820115 20920257 23761445 21719571 25244985 20966253 12364791 14747013 17210077 25186727 28797301 26830274 25576852 28049606 19178717 25315136 22216098 24508170
27762356 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 12110170 12660785 20431018 27129103 17510324 26369729 20798317 24845553 24945155 15632085 27966611 25401762 26823975 20566863 21347285 18362917 19820115 20920257 23761445 21719571 25244985 20966253 12364791 14747013 17210077 25186727 28797301 26830274 25576852 28049606 19178717 25315136 22216098 24508170
EMBL
DQ311302
ABD36246.1
GDQN01004555
JAT86499.1
KQ459593
KPI96321.1
+ More
ODYU01002980 SOQ41161.1 KZ149962 PZC76292.1 RSAL01000024 RVE52238.1 GAIX01006054 JAA86506.1 KQ460152 KPJ17419.1 NWSH01000104 PCG79521.1 AGBW02011569 OWR46479.1 CH940648 EDW62197.1 CH902619 EDV36054.1 CH964282 EDW85758.1 CH933808 EDW08735.1 CH916367 EDW01841.1 CH954179 EDV55050.1 CH982321 CM002911 EDX15593.1 KMY94796.1 GFDL01000965 JAV34080.1 BC082697 BC106244 CM004479 AAH82697.1 AAI06245.1 OCT71113.1 CM000362 EDX07644.1 AE013599 AHN56361.1 CM000158 EDW91660.1 AF512426 BT030940 BT030980 AAMC01106331 AAMC01106332 AAMC01106333 BC121544 CR761406 KV460372 AAI21545.1 CAJ82298.1 OCA19673.1 GDKW01001491 JAI55104.1 ACPB03000917 GAHY01001452 JAA76058.1 BC087298 CM004478 AAH87298.1 OCT73485.1 CH477367 EAT42511.1 CH480816 EDW48430.1 GANO01001117 JAB58754.1 GDAI01001557 JAI16046.1 GL449769 EFN82028.1 KK853026 KDR12309.1 GDHF01009370 JAI42944.1 GAPW01006433 GAPW01006432 GAPW01006431 GAPW01006430 JAC07168.1 GBYB01005797 JAG75564.1 GFTR01001584 JAW14842.1 GECL01001607 JAP04517.1 CM000071 EDY69212.1 CH479183 EDW36373.1 KZ021941 PIO13671.1 MG213852 AXY96274.1 OUUW01000001 SPP75890.1 KY031988 APR62714.1 GBHO01044986 GDHC01002115 JAF98617.1 JAQ16514.1 DS235271 EEB14161.1 KZ288220 PBC32149.1 JR048367 AEY60643.1 GECU01012196 JAS95510.1 KC741153 AGM32977.1 GECZ01018122 JAS51647.1 AXCM01005370 ADTU01018740 KQ971345 KYB27078.1 AXCN02000768 GBBK01004671 JAC19811.1 NEVH01006578 PNF37725.1 ADMH02001549 ETN62102.1 KQ978881 KYN27795.1 GL887908 EGI69475.1 GGFM01008487 MBW29238.1 KQ978406 KYM94168.1 AAAB01008964 EAA12574.2 GGFJ01011036 MBW60177.1 JXJN01020015 GFPF01010775 MAA21921.1 GEDV01010706 JAP77851.1 GACK01006535 JAA58499.1 GEFH01002709 JAP65872.1 EU934350 ACI30128.1 GGFK01011946 MBW45267.1 GFDF01011226 JAV02858.1 GEBQ01008900 JAT31077.1 GBBM01003424 JAC31994.1 JO843308 AEO34925.1 KK107021 EZA62338.1
ODYU01002980 SOQ41161.1 KZ149962 PZC76292.1 RSAL01000024 RVE52238.1 GAIX01006054 JAA86506.1 KQ460152 KPJ17419.1 NWSH01000104 PCG79521.1 AGBW02011569 OWR46479.1 CH940648 EDW62197.1 CH902619 EDV36054.1 CH964282 EDW85758.1 CH933808 EDW08735.1 CH916367 EDW01841.1 CH954179 EDV55050.1 CH982321 CM002911 EDX15593.1 KMY94796.1 GFDL01000965 JAV34080.1 BC082697 BC106244 CM004479 AAH82697.1 AAI06245.1 OCT71113.1 CM000362 EDX07644.1 AE013599 AHN56361.1 CM000158 EDW91660.1 AF512426 BT030940 BT030980 AAMC01106331 AAMC01106332 AAMC01106333 BC121544 CR761406 KV460372 AAI21545.1 CAJ82298.1 OCA19673.1 GDKW01001491 JAI55104.1 ACPB03000917 GAHY01001452 JAA76058.1 BC087298 CM004478 AAH87298.1 OCT73485.1 CH477367 EAT42511.1 CH480816 EDW48430.1 GANO01001117 JAB58754.1 GDAI01001557 JAI16046.1 GL449769 EFN82028.1 KK853026 KDR12309.1 GDHF01009370 JAI42944.1 GAPW01006433 GAPW01006432 GAPW01006431 GAPW01006430 JAC07168.1 GBYB01005797 JAG75564.1 GFTR01001584 JAW14842.1 GECL01001607 JAP04517.1 CM000071 EDY69212.1 CH479183 EDW36373.1 KZ021941 PIO13671.1 MG213852 AXY96274.1 OUUW01000001 SPP75890.1 KY031988 APR62714.1 GBHO01044986 GDHC01002115 JAF98617.1 JAQ16514.1 DS235271 EEB14161.1 KZ288220 PBC32149.1 JR048367 AEY60643.1 GECU01012196 JAS95510.1 KC741153 AGM32977.1 GECZ01018122 JAS51647.1 AXCM01005370 ADTU01018740 KQ971345 KYB27078.1 AXCN02000768 GBBK01004671 JAC19811.1 NEVH01006578 PNF37725.1 ADMH02001549 ETN62102.1 KQ978881 KYN27795.1 GL887908 EGI69475.1 GGFM01008487 MBW29238.1 KQ978406 KYM94168.1 AAAB01008964 EAA12574.2 GGFJ01011036 MBW60177.1 JXJN01020015 GFPF01010775 MAA21921.1 GEDV01010706 JAP77851.1 GACK01006535 JAA58499.1 GEFH01002709 JAP65872.1 EU934350 ACI30128.1 GGFK01011946 MBW45267.1 GFDF01011226 JAV02858.1 GEBQ01008900 JAT31077.1 GBBM01003424 JAC31994.1 JO843308 AEO34925.1 KK107021 EZA62338.1
Proteomes
UP000053268
UP000283053
UP000053240
UP000218220
UP000007151
UP000008792
+ More
UP000007801 UP000192221 UP000007798 UP000009192 UP000001070 UP000008711 UP000000304 UP000186698 UP000000803 UP000002282 UP000008143 UP000015103 UP000008820 UP000001292 UP000008237 UP000027135 UP000001819 UP000008744 UP000268350 UP000005203 UP000009046 UP000242457 UP000075883 UP000005205 UP000007266 UP000075881 UP000075886 UP000075884 UP000235965 UP000091820 UP000000673 UP000078492 UP000007755 UP000075901 UP000076408 UP000078542 UP000075903 UP000075902 UP000075882 UP000007062 UP000075900 UP000075920 UP000092460 UP000261460 UP000264840 UP000261580 UP000265100 UP000265160 UP000075885 UP000075880 UP000092443 UP000261640 UP000069272 UP000095301 UP000053097 UP000265040
UP000007801 UP000192221 UP000007798 UP000009192 UP000001070 UP000008711 UP000000304 UP000186698 UP000000803 UP000002282 UP000008143 UP000015103 UP000008820 UP000001292 UP000008237 UP000027135 UP000001819 UP000008744 UP000268350 UP000005203 UP000009046 UP000242457 UP000075883 UP000005205 UP000007266 UP000075881 UP000075886 UP000075884 UP000235965 UP000091820 UP000000673 UP000078492 UP000007755 UP000075901 UP000076408 UP000078542 UP000075903 UP000075902 UP000075882 UP000007062 UP000075900 UP000075920 UP000092460 UP000261460 UP000264840 UP000261580 UP000265100 UP000265160 UP000075885 UP000075880 UP000092443 UP000261640 UP000069272 UP000095301 UP000053097 UP000265040
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
Q2F5X1
A0A1E1WHM4
A0A194PSA5
A0A2H1VK38
A0A2W1BMJ0
A0A3S2M6R6
+ More
S4P4B3 A0A194RJ04 A0A2A4K5L7 A0A212EYA0 B4LNL7 B3MBC9 A0A1W4UJV8 B4NN19 B4KM71 B4J5R5 B3NMP6 B4NSL5 A0A1Q3G2Q4 Q640D2 B4QCC0 A0A0B4LFM5 B4P4N7 Q86BE9 Q28GF7 A0A0N7Z955 R4G877 Q5PQA0 Q178A8 B4HNL1 U5EVA9 A0A0K8TPH9 E2BQH0 A0A067QRX9 A0A0K8VVP3 A0A023ECS1 A0A0C9QYR3 A0A224Y1G1 A0A0V0GB54 B5DZQ9 B4GIQ3 A0A2G9QEV9 A0A385JF77 A0A3B0JKK7 A0A1X9IXE3 A0A0A9VWP5 A0A088AD44 E0VLA5 A0A2A3ELV5 V9IHQ6 A0A1B6J8L5 R4V2K3 A0A1B6FN65 A0A182MW29 A0A158NK86 A0A139WGF1 A0A182JU72 A0A182QE36 A0A023FD68 A0A182NUB4 A0A2J7RA64 A0A1A9WJE2 W5JCM5 A0A195EHR5 F4W8I4 A0A182SWV9 A0A2M3ZLE9 A0A182Y057 A0A151I7U1 A0A182VK82 A0A182TTD2 A0A182L1J5 Q7Q3Y1 A0A2M4C4C1 A0A182RZT2 A0A182W754 A0A1B0BT68 A0A3B4G386 A0A3Q2VAI7 A0A3Q4HTN4 A0A3P8N743 A0A3P9C1U2 A0A224Z4M2 A0A131YIU7 L7M5X3 A0A131XHN3 A0A182PMN5 A0A182IKZ2 A0A1A9YG60 B6DDY1 A0A3Q3MLH6 A0A2M4AWT9 A0A182F3E4 A0A1I8N311 A0A1L8D8T5 A0A1B6M571 A0A023GDT2 G3MN57 A0A026X2M0 A0A3Q1HYZ0
S4P4B3 A0A194RJ04 A0A2A4K5L7 A0A212EYA0 B4LNL7 B3MBC9 A0A1W4UJV8 B4NN19 B4KM71 B4J5R5 B3NMP6 B4NSL5 A0A1Q3G2Q4 Q640D2 B4QCC0 A0A0B4LFM5 B4P4N7 Q86BE9 Q28GF7 A0A0N7Z955 R4G877 Q5PQA0 Q178A8 B4HNL1 U5EVA9 A0A0K8TPH9 E2BQH0 A0A067QRX9 A0A0K8VVP3 A0A023ECS1 A0A0C9QYR3 A0A224Y1G1 A0A0V0GB54 B5DZQ9 B4GIQ3 A0A2G9QEV9 A0A385JF77 A0A3B0JKK7 A0A1X9IXE3 A0A0A9VWP5 A0A088AD44 E0VLA5 A0A2A3ELV5 V9IHQ6 A0A1B6J8L5 R4V2K3 A0A1B6FN65 A0A182MW29 A0A158NK86 A0A139WGF1 A0A182JU72 A0A182QE36 A0A023FD68 A0A182NUB4 A0A2J7RA64 A0A1A9WJE2 W5JCM5 A0A195EHR5 F4W8I4 A0A182SWV9 A0A2M3ZLE9 A0A182Y057 A0A151I7U1 A0A182VK82 A0A182TTD2 A0A182L1J5 Q7Q3Y1 A0A2M4C4C1 A0A182RZT2 A0A182W754 A0A1B0BT68 A0A3B4G386 A0A3Q2VAI7 A0A3Q4HTN4 A0A3P8N743 A0A3P9C1U2 A0A224Z4M2 A0A131YIU7 L7M5X3 A0A131XHN3 A0A182PMN5 A0A182IKZ2 A0A1A9YG60 B6DDY1 A0A3Q3MLH6 A0A2M4AWT9 A0A182F3E4 A0A1I8N311 A0A1L8D8T5 A0A1B6M571 A0A023GDT2 G3MN57 A0A026X2M0 A0A3Q1HYZ0
PDB
6IYC
E-value=1.94056e-23,
Score=263
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
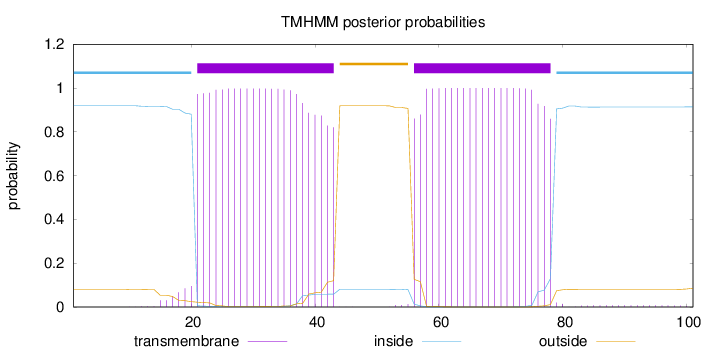
Length:
101
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.03898
Exp number, first 60 AAs:
27.18877
Total prob of N-in:
0.91902
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 55
TMhelix
56 - 78
inside
79 - 101
Population Genetic Test Statistics
Pi
188.576937
Theta
194.805054
Tajima's D
0.789827
CLR
1.11934
CSRT
0.603869806509674
Interpretation
Uncertain
