Gene
KWMTBOMO14051 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011234
Annotation
PREDICTED:_uncharacterized_protein_LOC106138920_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 3.426
Sequence
CDS
ATGCCATTTCAGAGGCCCGCCAGTGTTCCCATCGGGCAGGTATGGAGTCGTTTTAAAGGCAAAGAGAGGGATGGAAAACCAGCTAAGATGTACCAAATAAGAGACATGGACGACGCGAAAGAGAAATGCTTAGAGTTAATGAAGAGTACTTTTTTGAGGGACGAGCCGCTTTGTGATGTATTAGAAATCAACAGGGATCCCGTTTCAATTGAGACTATAATGAAAAACTGGGAGGAATACGTAGCGCAGAACGTCAGTCTGGCTTGTTACACCGAGGTAGACGGGGAACCTGACGAGCTGGTCGGCTTCAACATAGTCCTGGTGAAGAGCATCGACGATATAGACGAGGATTTAGATACGGTAAAAGGCGAATCCTGGAAGAAGCTGCTTAGGACACTGATAGAAGCGGAGAATTTGTGCAACGTATTCAAATACTACGGCGTGGACAAGTACTTGACCTCGAGCGGGCTCACGGTGCTGCCCGAGCACCGAGGTCAGAACATCGGAGCCAGACTCTTCGCGGCCAGAGAGCCCCTGGGAAACGCTCTTGGCATAGAGGCGACTGCTACCGTCTTCACGGCCACCACGTCCCAGGTCTTGGCAGCGAAGTGTGGCTACGAGACGTTAGCGGAACTAAAATACTCAGACATGTTAAAGTTCGGAATAAATCTGACTGAATGCACCACAGACAGAGCAAAACTAATGGGCACTCGATTCAAAAAACAACAATAA
Protein
MPFQRPASVPIGQVWSRFKGKERDGKPAKMYQIRDMDDAKEKCLELMKSTFLRDEPLCDVLEINRDPVSIETIMKNWEEYVAQNVSLACYTEVDGEPDELVGFNIVLVKSIDDIDEDLDTVKGESWKKLLRTLIEAENLCNVFKYYGVDKYLTSSGLTVLPEHRGQNIGARLFAAREPLGNALGIEATATVFTATTSQVLAAKCGYETLAELKYSDMLKFGINLTECTTDRAKLMGTRFKKQQ
Summary
Uniprot
H9JNY0
A0A2W1BLY5
A0A2A4JY09
A0A3S2PHF9
A0A194PSQ5
A0A194RIK5
+ More
A0A212EK92 A0A0L7LBW0 A0A182H6Q5 A0A023EKP0 A0A023EKN3 A0A336LW64 Q1HQL2 A0A1S4FXC1 Q16KV2 A0A1Q3FLZ5 A0A023EKE6 A0A336LMK9 A0A182HYS0 R4FLD1 A0A182XIF8 A0A182KZH2 Q7PQC6 A0A2P8XJF0 A0A182RE13 A0A2H1WR66 A0A182W3I8 A0A182VIU0 A0A2A4JP93 B0W0U0 A0A182LT41 A0A182K8G4 U5EV53 A0A182TCZ0 A0A182T8F4 A0A182Y8P3 A0A084WTX4 A0A3B0EUA9 A0A182W3I9 A0A088ARM7 A0A182LT43 A0A194RJ24 A0A2P8XJC4 A0A182XIF9 A0A182KZH0 A0A182QBT5 Q7Q9Y6 A0A182Y8Q0 A0A182HYR9 A0A182J1P0 A0A182N9W9 A0A182SM53 A0A212EWK4 A0A182V961 A0A182PHW6 A0A182U689 S4NIK2 A0A182K8G5 A0A182G4I2 A0A182H6Q0 A0A182RE14 A0A2P8Y2T7 A0A182N9X0 U5ENP2 A0A1L8DAT3 A0A182PHW7 A0A2M4AYK9 Q1HR96 A0A2M4AR94 U5EMM4 B0W0T9 A0A1Y1NH41 A0A336LZ29 A0A1Y1L937 A0A1Y1L8Z0 A0A2P8XF55 A0A0N1PH01 A0A2M4AJL0 A0A2A3ELM2 B0W0U1 A0A1S4FXQ0 E0W1H0 A0A1L8DD29 A0A1L8DAF7 A0A084WTX5 A0A182H6Q3 Q16KU8 A0A1Y1MRC9 I4DLB4 A0A1B0GHD7 A0A0A9WE96 A0A0N0BJ59 A0A2M4BY13 Q16KV1 A0A1W4W8S3 A0A194Q6V7 A0A182F917
A0A212EK92 A0A0L7LBW0 A0A182H6Q5 A0A023EKP0 A0A023EKN3 A0A336LW64 Q1HQL2 A0A1S4FXC1 Q16KV2 A0A1Q3FLZ5 A0A023EKE6 A0A336LMK9 A0A182HYS0 R4FLD1 A0A182XIF8 A0A182KZH2 Q7PQC6 A0A2P8XJF0 A0A182RE13 A0A2H1WR66 A0A182W3I8 A0A182VIU0 A0A2A4JP93 B0W0U0 A0A182LT41 A0A182K8G4 U5EV53 A0A182TCZ0 A0A182T8F4 A0A182Y8P3 A0A084WTX4 A0A3B0EUA9 A0A182W3I9 A0A088ARM7 A0A182LT43 A0A194RJ24 A0A2P8XJC4 A0A182XIF9 A0A182KZH0 A0A182QBT5 Q7Q9Y6 A0A182Y8Q0 A0A182HYR9 A0A182J1P0 A0A182N9W9 A0A182SM53 A0A212EWK4 A0A182V961 A0A182PHW6 A0A182U689 S4NIK2 A0A182K8G5 A0A182G4I2 A0A182H6Q0 A0A182RE14 A0A2P8Y2T7 A0A182N9X0 U5ENP2 A0A1L8DAT3 A0A182PHW7 A0A2M4AYK9 Q1HR96 A0A2M4AR94 U5EMM4 B0W0T9 A0A1Y1NH41 A0A336LZ29 A0A1Y1L937 A0A1Y1L8Z0 A0A2P8XF55 A0A0N1PH01 A0A2M4AJL0 A0A2A3ELM2 B0W0U1 A0A1S4FXQ0 E0W1H0 A0A1L8DD29 A0A1L8DAF7 A0A084WTX5 A0A182H6Q3 Q16KU8 A0A1Y1MRC9 I4DLB4 A0A1B0GHD7 A0A0A9WE96 A0A0N0BJ59 A0A2M4BY13 Q16KV1 A0A1W4W8S3 A0A194Q6V7 A0A182F917
Pubmed
EMBL
BABH01009767
BABH01009768
KZ149971
PZC76069.1
NWSH01000403
PCG76656.1
+ More
RSAL01000031 RVE51573.1 KQ459593 KPI96342.1 KQ460152 KPJ17392.1 AGBW02014309 OWR41901.1 JTDY01001867 KOB72686.1 JXUM01114724 JXUM01114725 KQ565812 KXJ70646.1 GAPW01003862 JAC09736.1 GAPW01003872 JAC09726.1 UFQT01000017 SSX17868.1 DQ440432 ABF18465.1 CH477937 EAT34934.1 GFDL01006493 JAV28552.1 GAPW01003983 JAC09615.1 SSX17869.1 APCN01005402 GAHY01002149 JAA75361.1 AAAB01008898 EAA09041.4 PYGN01001937 PSN32094.1 ODYU01010443 SOQ55559.1 NWSH01000987 PCG73230.1 DS231819 EDS42757.1 AXCM01000548 GANO01003565 JAB56306.1 ATLV01026931 KE525420 KFB53668.1 RBVL01000169 RKO11195.1 KPJ17439.1 PSN32096.1 AXCN02001202 EAA09295.2 AGBW02011952 OWR45872.1 GAIX01014039 JAA78521.1 JXUM01042501 KQ561327 KXJ78967.1 JXUM01114723 KXJ70641.1 PYGN01001003 PSN38573.1 GANO01004974 JAB54897.1 GFDF01010634 JAV03450.1 GGFK01012558 MBW45879.1 DQ440198 ABF18231.1 GGFK01009995 MBW43316.1 GANO01004420 JAB55451.1 EDS42756.1 GEZM01002705 JAV97161.1 UFQS01000047 UFQT01000047 SSW98552.1 SSX18938.1 GEZM01063098 JAV69311.1 GEZM01063101 JAV69308.1 PYGN01002387 PSN30584.1 KQ460419 KPJ14861.1 GGFK01007654 MBW40975.1 KZ288222 PBC32096.1 EDS42758.1 DS235870 EEB19476.1 GFDF01009807 JAV04277.1 GFDF01010628 JAV03456.1 KFB53669.1 KXJ70644.1 EAT34938.1 GEZM01023729 JAV88211.1 AK402082 BAM18704.1 AJWK01004477 GBHO01037893 GBRD01005077 GBRD01005075 GDHC01015779 GDHC01001169 JAG05711.1 JAG60744.1 JAQ02850.1 JAQ17460.1 KQ435720 KOX78638.1 GGFJ01008808 MBW57949.1 EAT34935.1 KQ459581 KPI99125.1
RSAL01000031 RVE51573.1 KQ459593 KPI96342.1 KQ460152 KPJ17392.1 AGBW02014309 OWR41901.1 JTDY01001867 KOB72686.1 JXUM01114724 JXUM01114725 KQ565812 KXJ70646.1 GAPW01003862 JAC09736.1 GAPW01003872 JAC09726.1 UFQT01000017 SSX17868.1 DQ440432 ABF18465.1 CH477937 EAT34934.1 GFDL01006493 JAV28552.1 GAPW01003983 JAC09615.1 SSX17869.1 APCN01005402 GAHY01002149 JAA75361.1 AAAB01008898 EAA09041.4 PYGN01001937 PSN32094.1 ODYU01010443 SOQ55559.1 NWSH01000987 PCG73230.1 DS231819 EDS42757.1 AXCM01000548 GANO01003565 JAB56306.1 ATLV01026931 KE525420 KFB53668.1 RBVL01000169 RKO11195.1 KPJ17439.1 PSN32096.1 AXCN02001202 EAA09295.2 AGBW02011952 OWR45872.1 GAIX01014039 JAA78521.1 JXUM01042501 KQ561327 KXJ78967.1 JXUM01114723 KXJ70641.1 PYGN01001003 PSN38573.1 GANO01004974 JAB54897.1 GFDF01010634 JAV03450.1 GGFK01012558 MBW45879.1 DQ440198 ABF18231.1 GGFK01009995 MBW43316.1 GANO01004420 JAB55451.1 EDS42756.1 GEZM01002705 JAV97161.1 UFQS01000047 UFQT01000047 SSW98552.1 SSX18938.1 GEZM01063098 JAV69311.1 GEZM01063101 JAV69308.1 PYGN01002387 PSN30584.1 KQ460419 KPJ14861.1 GGFK01007654 MBW40975.1 KZ288222 PBC32096.1 EDS42758.1 DS235870 EEB19476.1 GFDF01009807 JAV04277.1 GFDF01010628 JAV03456.1 KFB53669.1 KXJ70644.1 EAT34938.1 GEZM01023729 JAV88211.1 AK402082 BAM18704.1 AJWK01004477 GBHO01037893 GBRD01005077 GBRD01005075 GDHC01015779 GDHC01001169 JAG05711.1 JAG60744.1 JAQ02850.1 JAQ17460.1 KQ435720 KOX78638.1 GGFJ01008808 MBW57949.1 EAT34935.1 KQ459581 KPI99125.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000069940 UP000249989 UP000008820 UP000075840 UP000076407 UP000075882 UP000007062 UP000245037 UP000075900 UP000075920 UP000075903 UP000002320 UP000075883 UP000075881 UP000075902 UP000075901 UP000076408 UP000030765 UP000276569 UP000005203 UP000075886 UP000075880 UP000075884 UP000075885 UP000242457 UP000009046 UP000092461 UP000053105 UP000192223 UP000069272
UP000037510 UP000069940 UP000249989 UP000008820 UP000075840 UP000076407 UP000075882 UP000007062 UP000245037 UP000075900 UP000075920 UP000075903 UP000002320 UP000075883 UP000075881 UP000075902 UP000075901 UP000076408 UP000030765 UP000276569 UP000005203 UP000075886 UP000075880 UP000075884 UP000075885 UP000242457 UP000009046 UP000092461 UP000053105 UP000192223 UP000069272
PRIDE
SUPFAM
SSF55729
SSF55729
ProteinModelPortal
H9JNY0
A0A2W1BLY5
A0A2A4JY09
A0A3S2PHF9
A0A194PSQ5
A0A194RIK5
+ More
A0A212EK92 A0A0L7LBW0 A0A182H6Q5 A0A023EKP0 A0A023EKN3 A0A336LW64 Q1HQL2 A0A1S4FXC1 Q16KV2 A0A1Q3FLZ5 A0A023EKE6 A0A336LMK9 A0A182HYS0 R4FLD1 A0A182XIF8 A0A182KZH2 Q7PQC6 A0A2P8XJF0 A0A182RE13 A0A2H1WR66 A0A182W3I8 A0A182VIU0 A0A2A4JP93 B0W0U0 A0A182LT41 A0A182K8G4 U5EV53 A0A182TCZ0 A0A182T8F4 A0A182Y8P3 A0A084WTX4 A0A3B0EUA9 A0A182W3I9 A0A088ARM7 A0A182LT43 A0A194RJ24 A0A2P8XJC4 A0A182XIF9 A0A182KZH0 A0A182QBT5 Q7Q9Y6 A0A182Y8Q0 A0A182HYR9 A0A182J1P0 A0A182N9W9 A0A182SM53 A0A212EWK4 A0A182V961 A0A182PHW6 A0A182U689 S4NIK2 A0A182K8G5 A0A182G4I2 A0A182H6Q0 A0A182RE14 A0A2P8Y2T7 A0A182N9X0 U5ENP2 A0A1L8DAT3 A0A182PHW7 A0A2M4AYK9 Q1HR96 A0A2M4AR94 U5EMM4 B0W0T9 A0A1Y1NH41 A0A336LZ29 A0A1Y1L937 A0A1Y1L8Z0 A0A2P8XF55 A0A0N1PH01 A0A2M4AJL0 A0A2A3ELM2 B0W0U1 A0A1S4FXQ0 E0W1H0 A0A1L8DD29 A0A1L8DAF7 A0A084WTX5 A0A182H6Q3 Q16KU8 A0A1Y1MRC9 I4DLB4 A0A1B0GHD7 A0A0A9WE96 A0A0N0BJ59 A0A2M4BY13 Q16KV1 A0A1W4W8S3 A0A194Q6V7 A0A182F917
A0A212EK92 A0A0L7LBW0 A0A182H6Q5 A0A023EKP0 A0A023EKN3 A0A336LW64 Q1HQL2 A0A1S4FXC1 Q16KV2 A0A1Q3FLZ5 A0A023EKE6 A0A336LMK9 A0A182HYS0 R4FLD1 A0A182XIF8 A0A182KZH2 Q7PQC6 A0A2P8XJF0 A0A182RE13 A0A2H1WR66 A0A182W3I8 A0A182VIU0 A0A2A4JP93 B0W0U0 A0A182LT41 A0A182K8G4 U5EV53 A0A182TCZ0 A0A182T8F4 A0A182Y8P3 A0A084WTX4 A0A3B0EUA9 A0A182W3I9 A0A088ARM7 A0A182LT43 A0A194RJ24 A0A2P8XJC4 A0A182XIF9 A0A182KZH0 A0A182QBT5 Q7Q9Y6 A0A182Y8Q0 A0A182HYR9 A0A182J1P0 A0A182N9W9 A0A182SM53 A0A212EWK4 A0A182V961 A0A182PHW6 A0A182U689 S4NIK2 A0A182K8G5 A0A182G4I2 A0A182H6Q0 A0A182RE14 A0A2P8Y2T7 A0A182N9X0 U5ENP2 A0A1L8DAT3 A0A182PHW7 A0A2M4AYK9 Q1HR96 A0A2M4AR94 U5EMM4 B0W0T9 A0A1Y1NH41 A0A336LZ29 A0A1Y1L937 A0A1Y1L8Z0 A0A2P8XF55 A0A0N1PH01 A0A2M4AJL0 A0A2A3ELM2 B0W0U1 A0A1S4FXQ0 E0W1H0 A0A1L8DD29 A0A1L8DAF7 A0A084WTX5 A0A182H6Q3 Q16KU8 A0A1Y1MRC9 I4DLB4 A0A1B0GHD7 A0A0A9WE96 A0A0N0BJ59 A0A2M4BY13 Q16KV1 A0A1W4W8S3 A0A194Q6V7 A0A182F917
PDB
4FD7
E-value=2.62424e-24,
Score=275
Ontologies
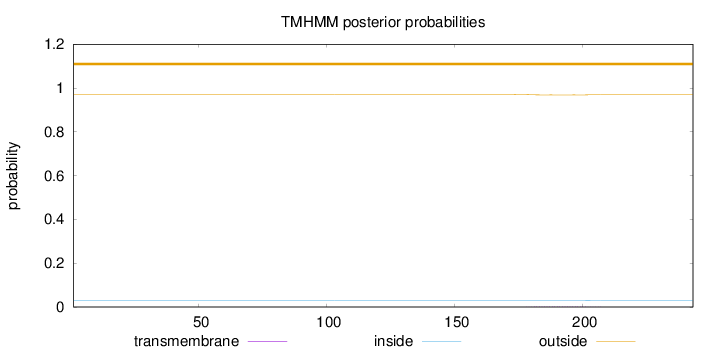
Topology
Length:
243
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04506
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02980
outside
1 - 243
Population Genetic Test Statistics
Pi
161.758769
Theta
141.406903
Tajima's D
0.417335
CLR
0.820111
CSRT
0.486525673716314
Interpretation
Uncertain
