Gene
KWMTBOMO14050 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011372
Annotation
glia_maturation_factor_beta_[Bombyx_mori]
Full name
Glia maturation factor
Location in the cell
Cytoplasmic Reliability : 2.354 Nuclear Reliability : 2.018
Sequence
CDS
ATGTCAGCACAAAACGTAAACGTCTGTGATATTGGTGATGATGTAAAGGAAGTCCTGAAAAAGTTTCGTTTCCAGAAACACACTTCCAATTCTGCTCTTATTTTAAAGGTGAATAGAGAAAAACAAGTCCTAGAAGTTGATGAGGAGCTGGAGAGTGTTGAGCTTGAAGAACTACAGGATATTCTGCCGTCCCATCAGCCGAGATTCATAGTTTACAGCTACAAAATGGAGCACAGTGACGGTCGGACGTCATTTCCGATGTGTTTTATATTCTACACACCCAGAGACGCACATATGGAGCTGCAAGTTATGTACGCGGGCACACAGAGGGCGCTGGCGGCCACCGTCGGAGCCCCGAGACTGCTCGAAGTCCGGGAGATAGACGAGCTGACCTCGGAATGGCTCGAAGAGAAACTGAGAAAATAA
Protein
MSAQNVNVCDIGDDVKEVLKKFRFQKHTSNSALILKVNREKQVLEVDEELESVELEELQDILPSHQPRFIVYSYKMEHSDGRTSFPMCFIFYTPRDAHMELQVMYAGTQRALAATVGAPRLLEVREIDELTSEWLEEKLRK
Summary
Similarity
Belongs to the actin-binding proteins ADF family. GMF subfamily.
Uniprot
H9JPB8
Q1HPS3
A0A2H1VRU8
A0A194PSC7
I4DQ53
A0A2W1BMA9
+ More
A0A1E1VZ42 A0A2A4JXC0 A0A212EK58 S4PRZ2 A0A2S2Q1T9 A0A2H8TUT9 A0A2S2NZN4 J9JYJ7 W8BTT5 A0A1B6LI11 A0A1B6CIG1 A0A0A1WK51 A0A0K8W601 A0A034VTR4 A0A1B0FH67 A0A232FHE8 A0A0A9YIS6 A0A1A9Y5B1 A0A1A9UR02 A0A1B0C031 A0A1B0A3X8 A0A1B6GAN8 K7J0R4 D3TRZ1 T1GBH6 A0A3B0JKY3 A0A0T6ASW8 A0A0M4E9E8 A0A161MRI4 T1P9A1 B4KI18 B4LTD8 Q29NM1 A0A0V0G5Z4 A0A1L8EE25 A0A1I8P0P8 B4I213 B4Q6Z7 Q9VJL6 B4GQM8 B4JAC1 B4NXP8 A0A1B0CNX4 T1DNC8 A0A2M4CTD6 A0A2M3ZB78 A0A2M4AWG5 A0A182F321 A0A1W4VJY0 B4MZY7 R4G3L4 T1IE60 A0A224Y0W6 A0A2M4C303 A0A2M4C3B7 A0A2R7W0W2 A0A1Q3FY14 B3N657 B3MJP2 R4WDL6 A0A069DP79 A0A1L8E3I1 U5EYB5 B0WLA3 A0A1B0CZG1 A0A182JLT5 D6WLE8 A0A182JXB2 A0A182P3D5 A0A182YMU0 A0A182NAQ2 A0A182TJN1 A0A182XKL2 A0A182KYN7 Q7PW80 A0A182HZ00 A0A182QN53 Q16I76 A0A023EG60 A0A1L8E3H4 A0A182RWG9 A0A182VSL3 A0A182V8J2 A0A182GLB8 A0A336K927 A0A0K8SUH9 E0VNZ9 A0A1W4X3H9 A0A336MIW9 A0A1Y1L5E9 A0A1W7RH63 J3S4F0 T1E6F9
A0A1E1VZ42 A0A2A4JXC0 A0A212EK58 S4PRZ2 A0A2S2Q1T9 A0A2H8TUT9 A0A2S2NZN4 J9JYJ7 W8BTT5 A0A1B6LI11 A0A1B6CIG1 A0A0A1WK51 A0A0K8W601 A0A034VTR4 A0A1B0FH67 A0A232FHE8 A0A0A9YIS6 A0A1A9Y5B1 A0A1A9UR02 A0A1B0C031 A0A1B0A3X8 A0A1B6GAN8 K7J0R4 D3TRZ1 T1GBH6 A0A3B0JKY3 A0A0T6ASW8 A0A0M4E9E8 A0A161MRI4 T1P9A1 B4KI18 B4LTD8 Q29NM1 A0A0V0G5Z4 A0A1L8EE25 A0A1I8P0P8 B4I213 B4Q6Z7 Q9VJL6 B4GQM8 B4JAC1 B4NXP8 A0A1B0CNX4 T1DNC8 A0A2M4CTD6 A0A2M3ZB78 A0A2M4AWG5 A0A182F321 A0A1W4VJY0 B4MZY7 R4G3L4 T1IE60 A0A224Y0W6 A0A2M4C303 A0A2M4C3B7 A0A2R7W0W2 A0A1Q3FY14 B3N657 B3MJP2 R4WDL6 A0A069DP79 A0A1L8E3I1 U5EYB5 B0WLA3 A0A1B0CZG1 A0A182JLT5 D6WLE8 A0A182JXB2 A0A182P3D5 A0A182YMU0 A0A182NAQ2 A0A182TJN1 A0A182XKL2 A0A182KYN7 Q7PW80 A0A182HZ00 A0A182QN53 Q16I76 A0A023EG60 A0A1L8E3H4 A0A182RWG9 A0A182VSL3 A0A182V8J2 A0A182GLB8 A0A336K927 A0A0K8SUH9 E0VNZ9 A0A1W4X3H9 A0A336MIW9 A0A1Y1L5E9 A0A1W7RH63 J3S4F0 T1E6F9
Pubmed
19121390
26354079
22651552
28756777
22118469
23622113
+ More
24495485 25830018 25348373 28648823 25401762 20075255 20353571 25315136 17994087 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 23691247 26334808 18362917 19820115 25244985 20966253 12364791 14747013 17210077 17510324 24945155 26483478 20566863 28004739 26358130 23025625 23758969 25727380
24495485 25830018 25348373 28648823 25401762 20075255 20353571 25315136 17994087 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 23691247 26334808 18362917 19820115 25244985 20966253 12364791 14747013 17210077 17510324 24945155 26483478 20566863 28004739 26358130 23025625 23758969 25727380
EMBL
BABH01009767
DQ443329
ABF51418.1
ODYU01004071
SOQ43565.1
KQ459593
+ More
KPI96341.1 AK403935 BAM20043.1 KZ149971 PZC76068.1 GDQN01011092 JAT79962.1 NWSH01000403 PCG76657.1 AGBW02014309 OWR41902.1 GAIX01014313 JAA78247.1 GGMS01002421 GGMS01008129 MBY71624.1 MBY77332.1 GFXV01005904 MBW17709.1 GGMR01009903 MBY22522.1 ABLF02027280 GAMC01001790 JAC04766.1 GEBQ01016753 JAT23224.1 GEDC01024173 JAS13125.1 GBXI01014863 JAC99428.1 GDHF01005686 JAI46628.1 GAKP01013762 JAC45190.1 CCAG010009633 NNAY01000195 OXU30102.1 GBHO01011540 JAG32064.1 JXJN01023460 GECZ01010279 JAS59490.1 EZ424193 ADD20469.1 CAQQ02393106 OUUW01000006 SPP81473.1 LJIG01022904 KRT78138.1 CP012523 ALC39752.1 GEMB01001550 JAS01609.1 KA645284 AFP59913.1 CH933807 EDW11300.2 CH940649 EDW63908.2 CH379059 EAL34197.3 GECL01002648 JAP03476.1 GFDG01001841 JAV16958.1 CH480820 EDW54570.1 CM000361 CM002910 EDX05205.1 KMY90493.1 AE014134 BT014645 KX531853 AAF53517.2 AAT27269.1 ANY27663.1 CH479187 EDW39900.1 CH916368 EDW03792.1 CM000157 EDW89673.1 AJWK01021065 GAMD01003000 JAA98590.1 GGFL01004434 MBW68612.1 GGFM01004969 MBW25720.1 GGFK01011816 MBW45137.1 CH963920 EDW77922.2 GAHY01002105 JAA75405.1 ACPB03017572 GFTR01002357 JAW14069.1 GGFJ01010533 MBW59674.1 GGFJ01010520 MBW59661.1 KK854227 PTY13352.1 GFDL01002642 JAV32403.1 CH954177 EDV58095.2 CH902620 EDV31381.1 AK417803 BAN21018.1 GBGD01003403 JAC85486.1 GFDF01000807 JAV13277.1 GANO01002103 JAB57768.1 DS231983 EDS30387.1 AJVK01020757 KQ971343 EFA04102.2 AAAB01008984 EAA14756.3 APCN01005836 AXCN02000665 CH478101 CH477434 EAT33968.1 EAT40982.1 EAT40983.1 GAPW01005788 GAPW01005787 JAC07810.1 GFDF01000806 JAV13278.1 JXUM01071490 JXUM01071491 KQ562667 KXJ75360.1 UFQS01000127 UFQT01000127 SSX00148.1 SSX20528.1 GBRD01008947 JAG56874.1 DS235354 EEB15105.1 UFQT01000637 SSX25998.1 GEZM01063920 JAV68909.1 GDAY02000944 JAV50483.1 JU174651 GBEX01001366 AFJ50177.1 JAI13194.1 GAAZ01000966 GBKC01001437 GBKD01000868 JAA96977.1 JAG44633.1 JAG46750.1
KPI96341.1 AK403935 BAM20043.1 KZ149971 PZC76068.1 GDQN01011092 JAT79962.1 NWSH01000403 PCG76657.1 AGBW02014309 OWR41902.1 GAIX01014313 JAA78247.1 GGMS01002421 GGMS01008129 MBY71624.1 MBY77332.1 GFXV01005904 MBW17709.1 GGMR01009903 MBY22522.1 ABLF02027280 GAMC01001790 JAC04766.1 GEBQ01016753 JAT23224.1 GEDC01024173 JAS13125.1 GBXI01014863 JAC99428.1 GDHF01005686 JAI46628.1 GAKP01013762 JAC45190.1 CCAG010009633 NNAY01000195 OXU30102.1 GBHO01011540 JAG32064.1 JXJN01023460 GECZ01010279 JAS59490.1 EZ424193 ADD20469.1 CAQQ02393106 OUUW01000006 SPP81473.1 LJIG01022904 KRT78138.1 CP012523 ALC39752.1 GEMB01001550 JAS01609.1 KA645284 AFP59913.1 CH933807 EDW11300.2 CH940649 EDW63908.2 CH379059 EAL34197.3 GECL01002648 JAP03476.1 GFDG01001841 JAV16958.1 CH480820 EDW54570.1 CM000361 CM002910 EDX05205.1 KMY90493.1 AE014134 BT014645 KX531853 AAF53517.2 AAT27269.1 ANY27663.1 CH479187 EDW39900.1 CH916368 EDW03792.1 CM000157 EDW89673.1 AJWK01021065 GAMD01003000 JAA98590.1 GGFL01004434 MBW68612.1 GGFM01004969 MBW25720.1 GGFK01011816 MBW45137.1 CH963920 EDW77922.2 GAHY01002105 JAA75405.1 ACPB03017572 GFTR01002357 JAW14069.1 GGFJ01010533 MBW59674.1 GGFJ01010520 MBW59661.1 KK854227 PTY13352.1 GFDL01002642 JAV32403.1 CH954177 EDV58095.2 CH902620 EDV31381.1 AK417803 BAN21018.1 GBGD01003403 JAC85486.1 GFDF01000807 JAV13277.1 GANO01002103 JAB57768.1 DS231983 EDS30387.1 AJVK01020757 KQ971343 EFA04102.2 AAAB01008984 EAA14756.3 APCN01005836 AXCN02000665 CH478101 CH477434 EAT33968.1 EAT40982.1 EAT40983.1 GAPW01005788 GAPW01005787 JAC07810.1 GFDF01000806 JAV13278.1 JXUM01071490 JXUM01071491 KQ562667 KXJ75360.1 UFQS01000127 UFQT01000127 SSX00148.1 SSX20528.1 GBRD01008947 JAG56874.1 DS235354 EEB15105.1 UFQT01000637 SSX25998.1 GEZM01063920 JAV68909.1 GDAY02000944 JAV50483.1 JU174651 GBEX01001366 AFJ50177.1 JAI13194.1 GAAZ01000966 GBKC01001437 GBKD01000868 JAA96977.1 JAG44633.1 JAG46750.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000007819
UP000092444
+ More
UP000215335 UP000092443 UP000078200 UP000092460 UP000092445 UP000002358 UP000015102 UP000268350 UP000092553 UP000095301 UP000009192 UP000008792 UP000001819 UP000095300 UP000001292 UP000000304 UP000000803 UP000008744 UP000001070 UP000002282 UP000092461 UP000069272 UP000192221 UP000007798 UP000015103 UP000008711 UP000007801 UP000002320 UP000092462 UP000075880 UP000007266 UP000075881 UP000075885 UP000076408 UP000075884 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075886 UP000008820 UP000075900 UP000075920 UP000075903 UP000069940 UP000249989 UP000009046 UP000192223
UP000215335 UP000092443 UP000078200 UP000092460 UP000092445 UP000002358 UP000015102 UP000268350 UP000092553 UP000095301 UP000009192 UP000008792 UP000001819 UP000095300 UP000001292 UP000000304 UP000000803 UP000008744 UP000001070 UP000002282 UP000092461 UP000069272 UP000192221 UP000007798 UP000015103 UP000008711 UP000007801 UP000002320 UP000092462 UP000075880 UP000007266 UP000075881 UP000075885 UP000076408 UP000075884 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075886 UP000008820 UP000075900 UP000075920 UP000075903 UP000069940 UP000249989 UP000009046 UP000192223
Interpro
Gene 3D
ProteinModelPortal
H9JPB8
Q1HPS3
A0A2H1VRU8
A0A194PSC7
I4DQ53
A0A2W1BMA9
+ More
A0A1E1VZ42 A0A2A4JXC0 A0A212EK58 S4PRZ2 A0A2S2Q1T9 A0A2H8TUT9 A0A2S2NZN4 J9JYJ7 W8BTT5 A0A1B6LI11 A0A1B6CIG1 A0A0A1WK51 A0A0K8W601 A0A034VTR4 A0A1B0FH67 A0A232FHE8 A0A0A9YIS6 A0A1A9Y5B1 A0A1A9UR02 A0A1B0C031 A0A1B0A3X8 A0A1B6GAN8 K7J0R4 D3TRZ1 T1GBH6 A0A3B0JKY3 A0A0T6ASW8 A0A0M4E9E8 A0A161MRI4 T1P9A1 B4KI18 B4LTD8 Q29NM1 A0A0V0G5Z4 A0A1L8EE25 A0A1I8P0P8 B4I213 B4Q6Z7 Q9VJL6 B4GQM8 B4JAC1 B4NXP8 A0A1B0CNX4 T1DNC8 A0A2M4CTD6 A0A2M3ZB78 A0A2M4AWG5 A0A182F321 A0A1W4VJY0 B4MZY7 R4G3L4 T1IE60 A0A224Y0W6 A0A2M4C303 A0A2M4C3B7 A0A2R7W0W2 A0A1Q3FY14 B3N657 B3MJP2 R4WDL6 A0A069DP79 A0A1L8E3I1 U5EYB5 B0WLA3 A0A1B0CZG1 A0A182JLT5 D6WLE8 A0A182JXB2 A0A182P3D5 A0A182YMU0 A0A182NAQ2 A0A182TJN1 A0A182XKL2 A0A182KYN7 Q7PW80 A0A182HZ00 A0A182QN53 Q16I76 A0A023EG60 A0A1L8E3H4 A0A182RWG9 A0A182VSL3 A0A182V8J2 A0A182GLB8 A0A336K927 A0A0K8SUH9 E0VNZ9 A0A1W4X3H9 A0A336MIW9 A0A1Y1L5E9 A0A1W7RH63 J3S4F0 T1E6F9
A0A1E1VZ42 A0A2A4JXC0 A0A212EK58 S4PRZ2 A0A2S2Q1T9 A0A2H8TUT9 A0A2S2NZN4 J9JYJ7 W8BTT5 A0A1B6LI11 A0A1B6CIG1 A0A0A1WK51 A0A0K8W601 A0A034VTR4 A0A1B0FH67 A0A232FHE8 A0A0A9YIS6 A0A1A9Y5B1 A0A1A9UR02 A0A1B0C031 A0A1B0A3X8 A0A1B6GAN8 K7J0R4 D3TRZ1 T1GBH6 A0A3B0JKY3 A0A0T6ASW8 A0A0M4E9E8 A0A161MRI4 T1P9A1 B4KI18 B4LTD8 Q29NM1 A0A0V0G5Z4 A0A1L8EE25 A0A1I8P0P8 B4I213 B4Q6Z7 Q9VJL6 B4GQM8 B4JAC1 B4NXP8 A0A1B0CNX4 T1DNC8 A0A2M4CTD6 A0A2M3ZB78 A0A2M4AWG5 A0A182F321 A0A1W4VJY0 B4MZY7 R4G3L4 T1IE60 A0A224Y0W6 A0A2M4C303 A0A2M4C3B7 A0A2R7W0W2 A0A1Q3FY14 B3N657 B3MJP2 R4WDL6 A0A069DP79 A0A1L8E3I1 U5EYB5 B0WLA3 A0A1B0CZG1 A0A182JLT5 D6WLE8 A0A182JXB2 A0A182P3D5 A0A182YMU0 A0A182NAQ2 A0A182TJN1 A0A182XKL2 A0A182KYN7 Q7PW80 A0A182HZ00 A0A182QN53 Q16I76 A0A023EG60 A0A1L8E3H4 A0A182RWG9 A0A182VSL3 A0A182V8J2 A0A182GLB8 A0A336K927 A0A0K8SUH9 E0VNZ9 A0A1W4X3H9 A0A336MIW9 A0A1Y1L5E9 A0A1W7RH63 J3S4F0 T1E6F9
PDB
1V6F
E-value=2.03767e-29,
Score=315
Ontologies
GO
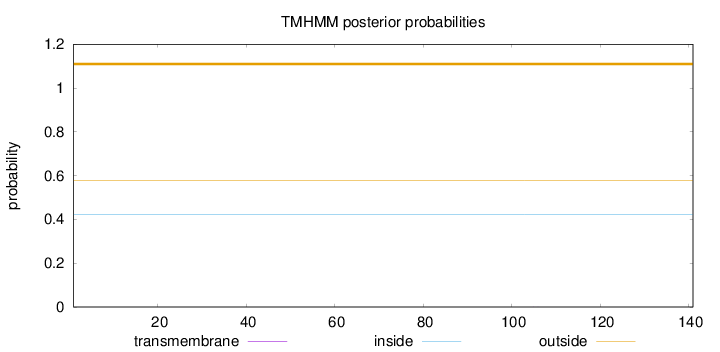
Topology
Length:
141
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00384
Exp number, first 60 AAs:
0
Total prob of N-in:
0.42188
outside
1 - 141
Population Genetic Test Statistics
Pi
180.292852
Theta
148.716553
Tajima's D
0.667424
CLR
1.099598
CSRT
0.566671666416679
Interpretation
Uncertain
