Gene
KWMTBOMO14047
Pre Gene Modal
BGIBMGA011235
Annotation
PREDICTED:_copper-transporting_ATPase_1_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.244
Sequence
CDS
ATGGCGGAGAATTTTAGCGACAATGATCAGGACCTGGTGGCGGTCAGGGTACCGATCAACGGTATGACATGCCAGTCGTGCGTCAGGAGCATCGAAGGATCGGTCAGAGACGTGCCAGGCGTGGAACGCGTTAAGGTGGAGTTATCAGAGAACGCCGGCTACATCAGATATGACCCCCGACTGTGTTCGCTGGAAGCGATCAGGTCTCACATAGAAAATATGGGGTTTGAGGTCCCGGTCGACAATGACGACGAAAGCAGAAATCTCCTGCCGAAGGATGTGCCGACCGCTATTATGATCGACATGACCGGCAGCCAGACGCCGCCCGTCAGCGAAGTCTTCCTCTCAGTGATCGGCATGACCTGCCAATCCTGCGTCAACACCATAGAAGGAGCGTTGAAAGCAAAGTCGGGAGTCACAAAGGGACACGTATCGCTCGCCGATGGAACGGCCCGCGTCGCCTTCCTGCCCAGCACCGTCACGCCCAGGGAGATAGCGGATCTGATATATGAACTGGGCTTCGACGTCAACATCATATCCGTCGACGGACAGGTTTATACGGAAAATAAGTCTCAAATATGTAACAACACAGAGGGATCTGGTGATTGCGCTAAGCAATTGTCTCCGTTCAAGAACAAACTTCAAACGAATGGTTCAGGTCCGCCGGTCGCCGAGGCGGAACTGTCCCGATGTACGCTGGAAGTGCGCGGGATGACCTGCGCCAGTTGTGTCTCTGCGATAGAAAAACATTGCACAAAATTATATGGGGTGCGTTCAGTGCTGATTGCTCTGTTGGCTGCGAAGGCCGAGGTCTGCTACGAGCCCCGTAAGATCTCGGCCTTAGACGTGGCCAATTCCATAACGGAGCTGGGCTTTCCGTCGGAGGTCATCAGCGAGAACGACGGCAGCGGCTTAAAGGAAATACATTTGCTGATAAAGGGAATGACGTGCGCTTCATGTGTCAATAAAATAGAGAAGACGGTACTTAAGCTGACCGGAGTCGAATCCTGCTCCGTGGCCCTGACGACATGCAAGGGCCGCGTCAAATACAACGCAGAACAGATCGGGCCCCGCACGATATGCGACGCGGTCGCCGGGCTCGGGTTCGAGGCGACGGTCGCCAGCCCCCACAACCGGGGGGAAACCAGCTATTTGGAGCACAGAGAGGAGATAAGGAAGTGGCGGAACGCGTTTCTGGTCTCGCTGGTGTTCGGGGCTCCGTGCATGGCCGCCATGGCGTATTTCATGTCCATGATGTCGCACGGCCACAGCGCCAAGGACATGTGCTGCGTGCTGCCCGGACTCAGTTTGGAGAACCTGGTCATGTTCCTGCTGGCGACGCCGGTGCAGTTCATAGGCGGCTGGCATTTCTACAAGCAAGCTTATAAAGCTCTAAAGCACGGCACGACGAACATGGACGTGCTGATAGCAATGACAACCACCATTAGCTACGTATATTCTCTAGGTGTGGTTGTAGCTTCCATGGCGACCCTCCAAGAGGCCAGTCCGGTCACGTTCTTCGACACGCCACCGATGTTGCTGGTCTTCGTATCTCTGGGCAGGTGGCTGGAGCACATAGCCAAGGGTAAAACATCGGAAGCCTTGTCGAAGCTGTTATCGCTGAAGGCAACGGAAGCCGTGCTGGTGACCCTGGACGACAGTGGTAACATTGTAAACGAGAAGTGCATACCGGTTGACCTGGTGGAACGCGGAGACATTCTGAAGGTAGTACCGGGCGCCAAGATACCGGTCGACGGCAAAGTCATATCCGGTCAGACGACGTGCGACGAGTCCCTGATAACCGGGGAATCTATGCCGGTGCCGAAGACGAAAGATTCCCTGGTGATCGGGGGCAGCATGAACTCCAACGGCGCCGTGCTGATGCTGGCGACCCACACCGGGGAAAGTTCGACCCTGGCCCAGATCGTGCGGCTGGTGGAGAGCGCGCAGAGCAGTAAGGCGCCCGTGCAACGACTCGCCGACACCATGGCGGGGTATTTCGTTCCGTTGGTGGTGTTCCTGTCGGTCCTGACGCTGGTCTGCTGGATCATAAGCGGAGCTCTGGACATAAACAGGATAAAGGCCATCACTCCTGAGGTGTACCTGAACGCTGGCTACTCGGACTGGGAGCTGATCGTGCAGTCGGCGTTCCACTTCGCGCTGTCGGTGCTCGCCATCGCGTGCCCGTGCGCGCTGGGGCTGGCCACGCCCACCGCCGTCATGGTGGCCACGGGCGTGGGCGCCAACCTGGGGCTGCTCATCAAGGGCGCGGAGCCGCTCGAGAACGCGCACAAGGTGAAAACTGTGATATTCGACAAGACCGGCACCATAACGCGGGGCTGCACCACCGTCGCCAGGCTGTGCCTCTTCACGGGGGAGCCGGCCGCCCTGCCGGACCTCGTCGCCTGCCTCCTCGCCGCCGAACTCAACTCCGAACACCCTGTAGCTTCAGCTATAGTCCAGTGGTGCAGTGCGGTGCTGAAGGGCGCGAACCCCCTGCAGTGTACCGGCTTCGTGGCCGCCCCTGGATGCGGCCTCAGGGCGAAGCTAAAAGCAACGAACAGCTCCGCCATACACGTGCCGGAGCTAGCCAACATCGTCAACCAGGCCAAGCCGGGGTGCACTTTCGACCTTCACGGCGTCCCCGTCGAGGTGGTGGCGGGCAGTGATGATGCCGCCCTGAGCTCGGCCCAGGCAGCCGCGAGACTACACCGATTGATCAGGACCGAGGATGGAGATGAGTCGGCGGAGCATATAGTCCTGATCGGGAACCGCGAGTGGATGAGTCGCAACGGAGTCAACGTGCCGCGGCCGGTCCAGAAGGCGCTGTCTCAACACGAGGAGCTCGGCTACACGGCCGTGCTGGCCGCCATCGATGATCAGCTGGTCTGCACCATCGGCGTCGCGGACGAAGTAAAGCCCGAAGCCCACCTGGCCGTCTACTGGCTGAAGAAGATGGGCCTCGAGGTGTGCCTGCTGACGGGAGACAACAGGAAGACAGCCGTCGCCATCGCCAGACAGGTCGGCATCAGGAAGGTGTACGCGGAGGTGCTGCCCTCGCACAAGGTCGCTAAAATACAAAAGCTGCAGGAGGAGGGGCAGAGGGTGGCCATGGTCGGTGACGGCGTCAACGATTCTCCAGCCCTGGCGCAGGCCGACGTGGGCATCGCCATCGCCAGCGGCACCGACGTAGCCGTGGAGGCCGCAGACCTGGTGCTCATGAGGAACGATCTTCTGGACGTGGTGGCCTGCTTGAACCTGTCCAGGACCACCGTGAGGAGGGTCAGGCTGAACTTCCTTTTCGCTTCCGTCTACAATCTGCTAGGGATACCGCTGGCGAGTGGAGCCTTCGCTTTATACGGAATGCAGCTGCAGCCCTGGATGGCGTCAGCTGCGATGGCGATGAGCTCAGTGTCCGTGGTCTGCTCAAGCTTGCTGCTCAAAACTTTCAGGAAACCAACGATCGAAGAGTTGAGGACGCCGGAGTACCTCCAGTCGATCCGGCTCGAGGATCTGGACAGCGTGTCCGTGCACCGGGGACTCGACGAGAGAGTGGACCCCAGCGACCGAAGTGTCACCGCTCTATCCAAATTTTTCAACAGAGCCAAAGCCGACGAGGGTTGCCTCCTGCAGGACGACGACGAGCTGGCCGCCGTCTCCTTCATACCGAAACAGCTTTCGCGCGACAGCCCTAGCCTGAACGCCTGA
Protein
MAENFSDNDQDLVAVRVPINGMTCQSCVRSIEGSVRDVPGVERVKVELSENAGYIRYDPRLCSLEAIRSHIENMGFEVPVDNDDESRNLLPKDVPTAIMIDMTGSQTPPVSEVFLSVIGMTCQSCVNTIEGALKAKSGVTKGHVSLADGTARVAFLPSTVTPREIADLIYELGFDVNIISVDGQVYTENKSQICNNTEGSGDCAKQLSPFKNKLQTNGSGPPVAEAELSRCTLEVRGMTCASCVSAIEKHCTKLYGVRSVLIALLAAKAEVCYEPRKISALDVANSITELGFPSEVISENDGSGLKEIHLLIKGMTCASCVNKIEKTVLKLTGVESCSVALTTCKGRVKYNAEQIGPRTICDAVAGLGFEATVASPHNRGETSYLEHREEIRKWRNAFLVSLVFGAPCMAAMAYFMSMMSHGHSAKDMCCVLPGLSLENLVMFLLATPVQFIGGWHFYKQAYKALKHGTTNMDVLIAMTTTISYVYSLGVVVASMATLQEASPVTFFDTPPMLLVFVSLGRWLEHIAKGKTSEALSKLLSLKATEAVLVTLDDSGNIVNEKCIPVDLVERGDILKVVPGAKIPVDGKVISGQTTCDESLITGESMPVPKTKDSLVIGGSMNSNGAVLMLATHTGESSTLAQIVRLVESAQSSKAPVQRLADTMAGYFVPLVVFLSVLTLVCWIISGALDINRIKAITPEVYLNAGYSDWELIVQSAFHFALSVLAIACPCALGLATPTAVMVATGVGANLGLLIKGAEPLENAHKVKTVIFDKTGTITRGCTTVARLCLFTGEPAALPDLVACLLAAELNSEHPVASAIVQWCSAVLKGANPLQCTGFVAAPGCGLRAKLKATNSSAIHVPELANIVNQAKPGCTFDLHGVPVEVVAGSDDAALSSAQAAARLHRLIRTEDGDESAEHIVLIGNREWMSRNGVNVPRPVQKALSQHEELGYTAVLAAIDDQLVCTIGVADEVKPEAHLAVYWLKKMGLEVCLLTGDNRKTAVAIARQVGIRKVYAEVLPSHKVAKIQKLQEEGQRVAMVGDGVNDSPALAQADVGIAIASGTDVAVEAADLVLMRNDLLDVVACLNLSRTTVRRVRLNFLFASVYNLLGIPLASGAFALYGMQLQPWMASAAMAMSSVSVVCSSLLLKTFRKPTIEELRTPEYLQSIRLEDLDSVSVHRGLDERVDPSDRSVTALSKFFNRAKADEGCLLQDDDELAAVSFIPKQLSRDSPSLNA
Summary
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IB subfamily.
Uniprot
H9JNY1
A0A3S2PID7
A0A212F1I0
A0A194PYV7
A0A088ARU7
A0A154PBY9
+ More
A0A2A3EM44 E2A8G0 A0A084WJ40 A0A0L7RDE4 A0A182N3K7 A0A336N3Y4 A0A336L514 A0A336MVP0 A0A1L8DEW1 A0A3L8DH34 A0A182PUK5 A0A1L8DET7 A0A1Q3FBB9 A0A182TEM7 A0A1Q3FDR7 A0A182L4N5 A0A182VAE4 A0A182HH64 A0A182IU19 A0A2M4CQI3 A0A1Q3FAQ5 A0A0J7L6F8 U5EGG8 E2C651 A0A182X6Q9 A0A1S4F4N4 A0A182H1I4 B0XIQ4 A0A182W1R0 A0A2M4A5K6 A0A2J7RI54 A0A2M4A5F9 B0WRZ5 A0A182PZZ9 W5JBD0 A0A2M4CQJ1 A0A182KCF6 A0A151XFI2 A0A182LZK5 A0A158NHF2 Q5TMM2 A0A195AZD5 A0A2M4BAL0 A0A182YL23 A0A2M4B9P2 A0A182RVE5 A0A310SRL6 A0A195D4L2 A0A2M3ZGJ4 A0A182FVU1 A0A195E2A2 Q17FH7 F4WD89 B4JMP4 A0A0L0CKX9 A0A195FWP6 A0A0K8U1J8 B4L6R5 A0A034VY80 B4NPT7 B4M7Q7 A0A0M4ERR1 A0A0C9RBJ3 A0A0N0BF76 B3MXJ1 A0A1B0GB32 B3NU80 A0A1W4WAJ7 A0A067R8T8 T1P9W3 A0A1I8MAC9 A0A0A1XNS8 W8APA1 V5GHJ7 B4Q1I5 Q6IDF6 Q9VYT4 A0A139WGD9 A0A139WG49 A0A1W4WLJ7 A0A1I8P499 B5DLH5 A0A1B6DN17 A0A1B6CFP2 A0A1B6DKB7 A0A1Y1MYS0 E0VL69 U4U8I3 A0A3B0K945 A0A2M4CZI3 A0A182G630 A0A3B0K6Z1 A0A2M4BB03 A0A069DXD5
A0A2A3EM44 E2A8G0 A0A084WJ40 A0A0L7RDE4 A0A182N3K7 A0A336N3Y4 A0A336L514 A0A336MVP0 A0A1L8DEW1 A0A3L8DH34 A0A182PUK5 A0A1L8DET7 A0A1Q3FBB9 A0A182TEM7 A0A1Q3FDR7 A0A182L4N5 A0A182VAE4 A0A182HH64 A0A182IU19 A0A2M4CQI3 A0A1Q3FAQ5 A0A0J7L6F8 U5EGG8 E2C651 A0A182X6Q9 A0A1S4F4N4 A0A182H1I4 B0XIQ4 A0A182W1R0 A0A2M4A5K6 A0A2J7RI54 A0A2M4A5F9 B0WRZ5 A0A182PZZ9 W5JBD0 A0A2M4CQJ1 A0A182KCF6 A0A151XFI2 A0A182LZK5 A0A158NHF2 Q5TMM2 A0A195AZD5 A0A2M4BAL0 A0A182YL23 A0A2M4B9P2 A0A182RVE5 A0A310SRL6 A0A195D4L2 A0A2M3ZGJ4 A0A182FVU1 A0A195E2A2 Q17FH7 F4WD89 B4JMP4 A0A0L0CKX9 A0A195FWP6 A0A0K8U1J8 B4L6R5 A0A034VY80 B4NPT7 B4M7Q7 A0A0M4ERR1 A0A0C9RBJ3 A0A0N0BF76 B3MXJ1 A0A1B0GB32 B3NU80 A0A1W4WAJ7 A0A067R8T8 T1P9W3 A0A1I8MAC9 A0A0A1XNS8 W8APA1 V5GHJ7 B4Q1I5 Q6IDF6 Q9VYT4 A0A139WGD9 A0A139WG49 A0A1W4WLJ7 A0A1I8P499 B5DLH5 A0A1B6DN17 A0A1B6CFP2 A0A1B6DKB7 A0A1Y1MYS0 E0VL69 U4U8I3 A0A3B0K945 A0A2M4CZI3 A0A182G630 A0A3B0K6Z1 A0A2M4BB03 A0A069DXD5
Pubmed
19121390
22118469
26354079
20798317
24438588
30249741
+ More
20966253 26483478 20920257 23761445 21347285 12364791 25244985 17510324 21719571 17994087 26108605 25348373 24845553 25315136 25830018 24495485 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 15632085 28004739 20566863 23537049 26334808
20966253 26483478 20920257 23761445 21347285 12364791 25244985 17510324 21719571 17994087 26108605 25348373 24845553 25315136 25830018 24495485 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 15632085 28004739 20566863 23537049 26334808
EMBL
BABH01009755
BABH01009756
RSAL01000024
RVE52226.1
AGBW02010873
OWR47592.1
+ More
KQ459593 KPI96340.1 KQ434856 KZC08730.1 KZ288222 PBC32111.1 GL437593 EFN70280.1 ATLV01023972 KE525347 KFB50234.1 KQ414614 KOC68858.1 UFQT01002308 SSX33188.1 UFQS01002004 UFQT01002004 SSX12928.1 SSX32370.1 SSX33189.1 GFDF01009106 JAV04978.1 QOIP01000008 RLU19737.1 GFDF01009111 JAV04973.1 GFDL01010198 JAV24847.1 GFDL01009339 JAV25706.1 APCN01002389 GGFL01003418 MBW67596.1 GFDL01010418 JAV24627.1 LBMM01000375 KMQ98953.1 GANO01003447 JAB56424.1 GL452926 EFN76571.1 JXUM01023160 KQ560653 KXJ81426.1 DS233353 EDS29576.1 GGFK01002587 MBW35908.1 NEVH01003505 PNF40517.1 GGFK01002725 MBW36046.1 DS232063 EDS33613.1 AXCN02001106 ADMH02001988 ETN60064.1 GGFL01003409 MBW67587.1 KQ982194 KYQ59095.1 AXCM01000696 ADTU01015732 ADTU01015733 ADTU01015734 AAAB01008986 EAL38875.3 KQ976694 KYM77603.1 GGFJ01000707 MBW49848.1 GGFJ01000633 MBW49774.1 KQ760869 OAD58764.1 KQ976870 KYN07781.1 GGFM01006888 MBW27639.1 KQ979763 KYN19206.1 CH477271 EAT45267.1 GL888086 EGI67816.1 CH916371 EDV91987.1 JRES01000262 KNC32887.1 KQ981208 KYN44851.1 GDHF01032109 JAI20205.1 CH933812 EDW06061.1 GAKP01012449 JAC46503.1 CH964291 EDW86527.2 CH940653 EDW62824.2 CP012528 ALC48865.1 GBYB01013879 GBYB01013880 JAG83646.1 JAG83647.1 KQ435814 KOX72672.1 CH902630 EDV38456.2 CCAG010006530 CCAG010006531 CH954180 EDV45995.2 KK852809 KDR16008.1 KA645556 AFP60185.1 GBXI01001690 JAD12602.1 GAMC01018709 GAMC01018708 JAB87847.1 GALX01004982 JAB63484.1 CM000162 EDX02474.2 BT014649 AAT27273.1 AE014298 AAF48104.3 AGB95309.1 KQ971348 KYB26921.1 KYB26922.1 CH379063 EDY72375.1 GEDC01010266 JAS27032.1 GEDC01025108 GEDC01024633 JAS12190.1 JAS12665.1 GEDC01011164 JAS26134.1 GEZM01017301 GEZM01017300 JAV90789.1 DS235271 EEB14125.1 KB632225 ERL90234.1 OUUW01000038 SPP89873.1 GGFL01006539 MBW70717.1 JXUM01044339 KQ561396 KXJ78716.1 SPP89874.1 GGFJ01001075 MBW50216.1 GBGD01000161 JAC88728.1
KQ459593 KPI96340.1 KQ434856 KZC08730.1 KZ288222 PBC32111.1 GL437593 EFN70280.1 ATLV01023972 KE525347 KFB50234.1 KQ414614 KOC68858.1 UFQT01002308 SSX33188.1 UFQS01002004 UFQT01002004 SSX12928.1 SSX32370.1 SSX33189.1 GFDF01009106 JAV04978.1 QOIP01000008 RLU19737.1 GFDF01009111 JAV04973.1 GFDL01010198 JAV24847.1 GFDL01009339 JAV25706.1 APCN01002389 GGFL01003418 MBW67596.1 GFDL01010418 JAV24627.1 LBMM01000375 KMQ98953.1 GANO01003447 JAB56424.1 GL452926 EFN76571.1 JXUM01023160 KQ560653 KXJ81426.1 DS233353 EDS29576.1 GGFK01002587 MBW35908.1 NEVH01003505 PNF40517.1 GGFK01002725 MBW36046.1 DS232063 EDS33613.1 AXCN02001106 ADMH02001988 ETN60064.1 GGFL01003409 MBW67587.1 KQ982194 KYQ59095.1 AXCM01000696 ADTU01015732 ADTU01015733 ADTU01015734 AAAB01008986 EAL38875.3 KQ976694 KYM77603.1 GGFJ01000707 MBW49848.1 GGFJ01000633 MBW49774.1 KQ760869 OAD58764.1 KQ976870 KYN07781.1 GGFM01006888 MBW27639.1 KQ979763 KYN19206.1 CH477271 EAT45267.1 GL888086 EGI67816.1 CH916371 EDV91987.1 JRES01000262 KNC32887.1 KQ981208 KYN44851.1 GDHF01032109 JAI20205.1 CH933812 EDW06061.1 GAKP01012449 JAC46503.1 CH964291 EDW86527.2 CH940653 EDW62824.2 CP012528 ALC48865.1 GBYB01013879 GBYB01013880 JAG83646.1 JAG83647.1 KQ435814 KOX72672.1 CH902630 EDV38456.2 CCAG010006530 CCAG010006531 CH954180 EDV45995.2 KK852809 KDR16008.1 KA645556 AFP60185.1 GBXI01001690 JAD12602.1 GAMC01018709 GAMC01018708 JAB87847.1 GALX01004982 JAB63484.1 CM000162 EDX02474.2 BT014649 AAT27273.1 AE014298 AAF48104.3 AGB95309.1 KQ971348 KYB26921.1 KYB26922.1 CH379063 EDY72375.1 GEDC01010266 JAS27032.1 GEDC01025108 GEDC01024633 JAS12190.1 JAS12665.1 GEDC01011164 JAS26134.1 GEZM01017301 GEZM01017300 JAV90789.1 DS235271 EEB14125.1 KB632225 ERL90234.1 OUUW01000038 SPP89873.1 GGFL01006539 MBW70717.1 JXUM01044339 KQ561396 KXJ78716.1 SPP89874.1 GGFJ01001075 MBW50216.1 GBGD01000161 JAC88728.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000005203
UP000076502
+ More
UP000242457 UP000000311 UP000030765 UP000053825 UP000075884 UP000279307 UP000075885 UP000075902 UP000075882 UP000075903 UP000075840 UP000075880 UP000036403 UP000008237 UP000076407 UP000069940 UP000249989 UP000002320 UP000075920 UP000235965 UP000075886 UP000000673 UP000075881 UP000075809 UP000075883 UP000005205 UP000007062 UP000078540 UP000076408 UP000075900 UP000078542 UP000069272 UP000078492 UP000008820 UP000007755 UP000001070 UP000037069 UP000078541 UP000009192 UP000007798 UP000008792 UP000092553 UP000053105 UP000007801 UP000092444 UP000008711 UP000192221 UP000027135 UP000095301 UP000002282 UP000000803 UP000007266 UP000192223 UP000095300 UP000001819 UP000009046 UP000030742 UP000268350
UP000242457 UP000000311 UP000030765 UP000053825 UP000075884 UP000279307 UP000075885 UP000075902 UP000075882 UP000075903 UP000075840 UP000075880 UP000036403 UP000008237 UP000076407 UP000069940 UP000249989 UP000002320 UP000075920 UP000235965 UP000075886 UP000000673 UP000075881 UP000075809 UP000075883 UP000005205 UP000007062 UP000078540 UP000076408 UP000075900 UP000078542 UP000069272 UP000078492 UP000008820 UP000007755 UP000001070 UP000037069 UP000078541 UP000009192 UP000007798 UP000008792 UP000092553 UP000053105 UP000007801 UP000092444 UP000008711 UP000192221 UP000027135 UP000095301 UP000002282 UP000000803 UP000007266 UP000192223 UP000095300 UP000001819 UP000009046 UP000030742 UP000268350
Pfam
PF00403 HMA
Interpro
IPR023214
HAD_sf
+ More
IPR036163 HMA_dom_sf
IPR036412 HAD-like_sf
IPR023299 ATPase_P-typ_cyto_dom_N
IPR017969 Heavy-metal-associated_CS
IPR001757 P_typ_ATPase
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR006122 HMA_Cu_ion-bd
IPR023298 ATPase_P-typ_TM_dom_sf
IPR006121 HMA_dom
IPR018303 ATPase_P-typ_P_site
IPR027256 P-typ_ATPase_IB
IPR036163 HMA_dom_sf
IPR036412 HAD-like_sf
IPR023299 ATPase_P-typ_cyto_dom_N
IPR017969 Heavy-metal-associated_CS
IPR001757 P_typ_ATPase
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR006122 HMA_Cu_ion-bd
IPR023298 ATPase_P-typ_TM_dom_sf
IPR006121 HMA_dom
IPR018303 ATPase_P-typ_P_site
IPR027256 P-typ_ATPase_IB
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9JNY1
A0A3S2PID7
A0A212F1I0
A0A194PYV7
A0A088ARU7
A0A154PBY9
+ More
A0A2A3EM44 E2A8G0 A0A084WJ40 A0A0L7RDE4 A0A182N3K7 A0A336N3Y4 A0A336L514 A0A336MVP0 A0A1L8DEW1 A0A3L8DH34 A0A182PUK5 A0A1L8DET7 A0A1Q3FBB9 A0A182TEM7 A0A1Q3FDR7 A0A182L4N5 A0A182VAE4 A0A182HH64 A0A182IU19 A0A2M4CQI3 A0A1Q3FAQ5 A0A0J7L6F8 U5EGG8 E2C651 A0A182X6Q9 A0A1S4F4N4 A0A182H1I4 B0XIQ4 A0A182W1R0 A0A2M4A5K6 A0A2J7RI54 A0A2M4A5F9 B0WRZ5 A0A182PZZ9 W5JBD0 A0A2M4CQJ1 A0A182KCF6 A0A151XFI2 A0A182LZK5 A0A158NHF2 Q5TMM2 A0A195AZD5 A0A2M4BAL0 A0A182YL23 A0A2M4B9P2 A0A182RVE5 A0A310SRL6 A0A195D4L2 A0A2M3ZGJ4 A0A182FVU1 A0A195E2A2 Q17FH7 F4WD89 B4JMP4 A0A0L0CKX9 A0A195FWP6 A0A0K8U1J8 B4L6R5 A0A034VY80 B4NPT7 B4M7Q7 A0A0M4ERR1 A0A0C9RBJ3 A0A0N0BF76 B3MXJ1 A0A1B0GB32 B3NU80 A0A1W4WAJ7 A0A067R8T8 T1P9W3 A0A1I8MAC9 A0A0A1XNS8 W8APA1 V5GHJ7 B4Q1I5 Q6IDF6 Q9VYT4 A0A139WGD9 A0A139WG49 A0A1W4WLJ7 A0A1I8P499 B5DLH5 A0A1B6DN17 A0A1B6CFP2 A0A1B6DKB7 A0A1Y1MYS0 E0VL69 U4U8I3 A0A3B0K945 A0A2M4CZI3 A0A182G630 A0A3B0K6Z1 A0A2M4BB03 A0A069DXD5
A0A2A3EM44 E2A8G0 A0A084WJ40 A0A0L7RDE4 A0A182N3K7 A0A336N3Y4 A0A336L514 A0A336MVP0 A0A1L8DEW1 A0A3L8DH34 A0A182PUK5 A0A1L8DET7 A0A1Q3FBB9 A0A182TEM7 A0A1Q3FDR7 A0A182L4N5 A0A182VAE4 A0A182HH64 A0A182IU19 A0A2M4CQI3 A0A1Q3FAQ5 A0A0J7L6F8 U5EGG8 E2C651 A0A182X6Q9 A0A1S4F4N4 A0A182H1I4 B0XIQ4 A0A182W1R0 A0A2M4A5K6 A0A2J7RI54 A0A2M4A5F9 B0WRZ5 A0A182PZZ9 W5JBD0 A0A2M4CQJ1 A0A182KCF6 A0A151XFI2 A0A182LZK5 A0A158NHF2 Q5TMM2 A0A195AZD5 A0A2M4BAL0 A0A182YL23 A0A2M4B9P2 A0A182RVE5 A0A310SRL6 A0A195D4L2 A0A2M3ZGJ4 A0A182FVU1 A0A195E2A2 Q17FH7 F4WD89 B4JMP4 A0A0L0CKX9 A0A195FWP6 A0A0K8U1J8 B4L6R5 A0A034VY80 B4NPT7 B4M7Q7 A0A0M4ERR1 A0A0C9RBJ3 A0A0N0BF76 B3MXJ1 A0A1B0GB32 B3NU80 A0A1W4WAJ7 A0A067R8T8 T1P9W3 A0A1I8MAC9 A0A0A1XNS8 W8APA1 V5GHJ7 B4Q1I5 Q6IDF6 Q9VYT4 A0A139WGD9 A0A139WG49 A0A1W4WLJ7 A0A1I8P499 B5DLH5 A0A1B6DN17 A0A1B6CFP2 A0A1B6DKB7 A0A1Y1MYS0 E0VL69 U4U8I3 A0A3B0K945 A0A2M4CZI3 A0A182G630 A0A3B0K6Z1 A0A2M4BB03 A0A069DXD5
PDB
3J09
E-value=9.56557e-65,
Score=631
Ontologies
GO
Topology
Subcellular location
Membrane
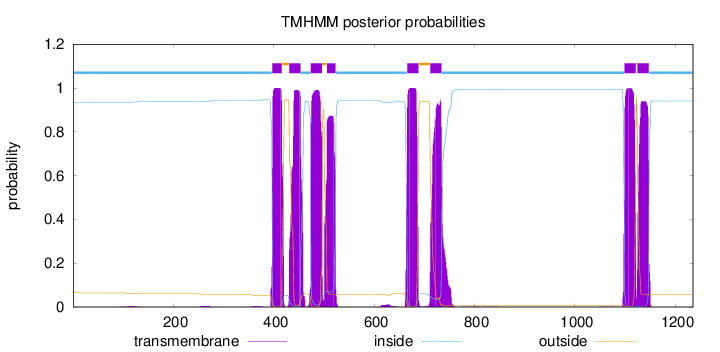
Length:
1235
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
171.81294
Exp number, first 60 AAs:
0.00138
Total prob of N-in:
0.93544
inside
1 - 396
TMhelix
397 - 416
outside
417 - 430
TMhelix
431 - 453
inside
454 - 473
TMhelix
474 - 496
outside
497 - 505
TMhelix
506 - 523
inside
524 - 665
TMhelix
666 - 688
outside
689 - 711
TMhelix
712 - 734
inside
735 - 1098
TMhelix
1099 - 1121
outside
1122 - 1124
TMhelix
1125 - 1147
inside
1148 - 1235
Population Genetic Test Statistics
Pi
218.944214
Theta
158.912566
Tajima's D
1.033064
CLR
0.370702
CSRT
0.670966451677416
Interpretation
Uncertain
