Gene
KWMTBOMO14043
Pre Gene Modal
BGIBMGA011369
Annotation
PREDICTED:_serine/threonine-protein_kinase_par-1-like_isoform_X3_[Bombyx_mori]
Full name
Non-specific serine/threonine protein kinase
Location in the cell
Nuclear Reliability : 2.156
Sequence
CDS
ATGGCCTGCAATATCAGCCCTATACACAAAAGCAAGCATTTCCCAATAGACCAGCTCGTGTGTGTCGGTAACTATGAACTTGAAAAGACTATAGGAACTGGTAACTTCGCCGTCGTAAAACTGGCTACACATGTTATTACTAAAAGTAAGGTCGCTATAAAAATAATTGACAAATCAAGATTAGATGAAGACAATTTGAAGAAGACATTCCGGGAAATAGCAATAATGAAGAGACTGAGTCATCCGCACATAGTTAGGTTATACCAGGTGATGGAGAGCACCCACACGATTTATTTAGTAACAGAATACGCACCGAACGGTGAAATATTTGATCACTTGGTGTCCAGGGGTCGTATGACGGAGTCGGAGGCGGCCCGAGCTTTCTCCCAGATGGTGGCCGCCGTGGGGTACTGCCACGCGAACGGTGTCGTCCATCGCGACCTAAAGGCTGAGAATTTACTGTTAGACAAAAACATGAACATAAAGTTAGCAGATTTCGGCTTCAGCAACGAATATAGAACCGGAGGAGCCCTGAGCACCTGGTGCGGGAGTCCTCCGTACGCAGCACCCGAGCTCTTCGAAGGGAGACAGTATGATGGACCCAAAGCCGACATTTGGTCACTGGGCGTCGTATTATACGTTCTGGTCTGCGGGGCGTTGCCGTTCGATGGGAACACCCTCCACGAGCTCAGGAACGTGGTCCTGAGCGGAAAGTTCCGAATACCGTACTTCATGTCACAAGAATGCGAAAATCTAATAAGGCATATGCTGGTCGTGGAACCGGAGAAGAGGTTATCCCTGCGCGGTGTTGCCAGGCACAGGTGGCTGGCGGCACACCAACCTGGGCCAGGACCGGGTGATCGAGATGCGTCGACCGGAGCCTGCATATGTCACACGTCTATGTCGCAGACCGAACCCGGGGCCTCGGAAACTGTTATGGCGCATTTGCTCACGCTCCCCGGGCTCACGAAGGAGCACGTACAACAGTCGGTGCAAGAAGAACGCTTCGATCACATATCGGCTATCTACCACCTCCTCATGGACAAGCTCGAACAGAGAAGCAGCGCCCGGAGTAGCCTGCAGCACAGAGATTCCCTGGACTCGACGACCGGTCTGCCGTTGGATCTCACTAGTTGTAAGAATTCGGAAGACGAGGAGATGGATGCGGAGGATTCTTCGATCCAGGATTCTCTGATATCGTTACCCAATCCTGAAGCGTCGGTGGACTACCTGACCGACGGACCCGAGGCTCTGGAGAAGTTCGGTGACGGCTCGATACCAGTTCAAGAGGAGCGGACCCAGCAGCCCGCCAGCACCGGCGAGCAGCACGCCACCAGGCGGCACACCGTGGGGCCCGGCGACTGCAGACACACACAGGGCGGCGAGCTGAGCGCGTGCGGGGCCGCGGTGTCCGCCGACCTGCGCCCCTCCCCCCTCTACGTATCCGCGCAGTTCAATGCCTCGCCGGACGTGTCGCTGCTGCCGAACACGAACCTAGCCGCCAAGCTGCCGGCCGTGCAGCACCAGCCCGCGCGCTACTTCAGCGTCAAGGACCAGCACCTGCTGAAGCCGCCGCACGCCATGCAGAGCCAAGGTACGAGCTACCTGCGGAGCTCCGTAAAGCAGTAA
Protein
MACNISPIHKSKHFPIDQLVCVGNYELEKTIGTGNFAVVKLATHVITKSKVAIKIIDKSRLDEDNLKKTFREIAIMKRLSHPHIVRLYQVMESTHTIYLVTEYAPNGEIFDHLVSRGRMTESEAARAFSQMVAAVGYCHANGVVHRDLKAENLLLDKNMNIKLADFGFSNEYRTGGALSTWCGSPPYAAPELFEGRQYDGPKADIWSLGVVLYVLVCGALPFDGNTLHELRNVVLSGKFRIPYFMSQECENLIRHMLVVEPEKRLSLRGVARHRWLAAHQPGPGPGDRDASTGACICHTSMSQTEPGASETVMAHLLTLPGLTKEHVQQSVQEERFDHISAIYHLLMDKLEQRSSARSSLQHRDSLDSTTGLPLDLTSCKNSEDEEMDAEDSSIQDSLISLPNPEASVDYLTDGPEALEKFGDGSIPVQEERTQQPASTGEQHATRRHTVGPGDCRHTQGGELSACGAAVSADLRPSPLYVSAQFNASPDVSLLPNTNLAAKLPAVQHQPARYFSVKDQHLLKPPHAMQSQGTSYLRSSVKQ
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
L-lysyl-[histone] + S-adenosyl-L-methionine = H(+) + N(6)-methyl-L-lysyl-[histone] + S-adenosyl-L-homocysteine
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
L-lysyl-[histone] + S-adenosyl-L-methionine = H(+) + N(6)-methyl-L-lysyl-[histone] + S-adenosyl-L-homocysteine
Uniprot
A0A2A4K7A3
A0A2W1BMK5
A0A2A4K5M6
A0A212EZB9
H9JPB6
A0A194RIZ9
+ More
A0A194PYU2 E0VPX8 K7ILZ4 A0A0L7QQB4 A0A0C9RV60 A0A0M9A067 A0A0C9QEA2 A0A151IQZ7 Q4QQA7 A0A232F6Z7 Q0IG70 A0A158NN80 A0A0A9Y3Q2 E2C476 A0A151HY62 A0A2A3E4E6 E2AQ24 A0A088APQ0 A0A224XLQ4 Q7Q5I9 A0A151X0P2 A0A195C3E7 A0NES6 B4HP02 A0A067R3B4 A0A026WRY8 A0A3L8DF00 A0A182YP73 A0A1J1HIR9 A0A182GE93 L7MMY1 A8DYI4 A0A336MWI8 A8DYI5 B4J7N2 A0A1B6H4M6 B3MBU0 L7LUJ6 A0A2S2NKQ2 A0A0B4LFW1 A1ZBC4 A0A023F273 A0A1S4G7H2 A0A0P8XMZ8 A0A0P8XMW6 A0A131XGE2 A0A131YQE6 A0A182QA15 B5DZ12 A0A0J9RFX1 A0A0J9U5M6 A0A084VXN3 B4P4A4 A0A0R1DS47 A0A0J9RG83 A0A0J9RHG4 A0A0R1DXK5 A0A0R1DRZ0 A0A2H8U050 A0A1L8DKS1 A0A154PEE2 L7LTW0 A0A1W4UA84 J9K8Y8 A0A1W4UNZ5 A0A1W4UA80 T1HPG7 A0A1E1X6R7 A0A0Q5VT30 B3NK99 B4KPL5 A0A1Y9IVD2 B4LJ49 A0A0Q5VKW8 A0A0Q5VLS1 A0A0Q9XKL0 A0A1I8Q3Z6 B4MJZ7 A0A0P5QAZ3 A0A1I8NCR8 A0A162RH65 A0A1I8NCR9 A0A1I8Q426 A0A0R3NS22 W8AKG4 A0A3B0JE14 A0A0R3NSH8 A0A0R3NLK6 T1JFK1 A0A0P4ZRN7 A0A0P5Y2V5
A0A194PYU2 E0VPX8 K7ILZ4 A0A0L7QQB4 A0A0C9RV60 A0A0M9A067 A0A0C9QEA2 A0A151IQZ7 Q4QQA7 A0A232F6Z7 Q0IG70 A0A158NN80 A0A0A9Y3Q2 E2C476 A0A151HY62 A0A2A3E4E6 E2AQ24 A0A088APQ0 A0A224XLQ4 Q7Q5I9 A0A151X0P2 A0A195C3E7 A0NES6 B4HP02 A0A067R3B4 A0A026WRY8 A0A3L8DF00 A0A182YP73 A0A1J1HIR9 A0A182GE93 L7MMY1 A8DYI4 A0A336MWI8 A8DYI5 B4J7N2 A0A1B6H4M6 B3MBU0 L7LUJ6 A0A2S2NKQ2 A0A0B4LFW1 A1ZBC4 A0A023F273 A0A1S4G7H2 A0A0P8XMZ8 A0A0P8XMW6 A0A131XGE2 A0A131YQE6 A0A182QA15 B5DZ12 A0A0J9RFX1 A0A0J9U5M6 A0A084VXN3 B4P4A4 A0A0R1DS47 A0A0J9RG83 A0A0J9RHG4 A0A0R1DXK5 A0A0R1DRZ0 A0A2H8U050 A0A1L8DKS1 A0A154PEE2 L7LTW0 A0A1W4UA84 J9K8Y8 A0A1W4UNZ5 A0A1W4UA80 T1HPG7 A0A1E1X6R7 A0A0Q5VT30 B3NK99 B4KPL5 A0A1Y9IVD2 B4LJ49 A0A0Q5VKW8 A0A0Q5VLS1 A0A0Q9XKL0 A0A1I8Q3Z6 B4MJZ7 A0A0P5QAZ3 A0A1I8NCR8 A0A162RH65 A0A1I8NCR9 A0A1I8Q426 A0A0R3NS22 W8AKG4 A0A3B0JE14 A0A0R3NSH8 A0A0R3NLK6 T1JFK1 A0A0P4ZRN7 A0A0P5Y2V5
EC Number
2.7.11.1
Pubmed
28756777
22118469
19121390
26354079
20566863
20075255
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 17510324 21347285 25401762 26823975 20798317 12364791 14747013 17210077 17994087 24845553 24508170 30249741 25244985 26483478 25576852 25474469 28049606 26830274 15632085 22936249 24438588 17550304 28503490 18057021 25315136 24495485
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 17510324 21347285 25401762 26823975 20798317 12364791 14747013 17210077 17994087 24845553 24508170 30249741 25244985 26483478 25576852 25474469 28049606 26830274 15632085 22936249 24438588 17550304 28503490 18057021 25315136 24495485
EMBL
NWSH01000104
PCG79532.1
KZ149962
PZC76289.1
PCG79531.1
AGBW02011345
+ More
OWR46801.1 BABH01009748 BABH01009749 BABH01009750 KQ460152 KPJ17414.1 KQ459593 KPI96325.1 DS235379 EEB15434.1 KQ414786 KOC60820.1 GBYB01012740 JAG82507.1 KQ435789 KOX74477.1 GBYB01012738 JAG82505.1 KQ981148 KYN09093.1 AE013599 BT023509 AAF57652.1 AAY84909.1 NNAY01000772 OXU26601.1 CH477261 EAT45687.1 ADTU01002507 ADTU01002508 ADTU01002509 ADTU01002510 ADTU01002511 ADTU01002512 ADTU01002513 ADTU01002514 ADTU01002515 ADTU01002516 GBHO01019479 GDHC01002131 JAG24125.1 JAQ16498.1 GL452426 EFN77247.1 KQ976747 KYM75320.1 KZ288391 PBC26364.1 GL441651 EFN64467.1 GFTR01007477 JAW08949.1 AAAB01008960 EAA11379.3 KQ982617 KYQ53809.1 KQ978350 KYM94703.1 EAU76597.1 CH480816 EDW48504.1 KK853026 KDR12311.1 KK107119 EZA58703.1 QOIP01000009 RLU18901.1 CVRI01000001 CRK86177.1 JXUM01009542 JXUM01009543 JXUM01009544 JXUM01010158 JXUM01010159 JXUM01010160 JXUM01010161 JXUM01010162 KQ560300 KQ560286 KXJ83174.1 KXJ83262.1 GACK01000070 JAA64964.1 ABV53845.2 UFQS01003371 UFQT01003371 SSX15571.1 SSX34934.1 ABV53846.2 CH916367 EDW02180.1 GECZ01000143 JAS69626.1 CH902619 EDV36111.2 GACK01009549 JAA55485.1 GGMR01005125 MBY17744.1 AHN56382.1 AAF57651.4 GBBI01003668 JAC15044.1 KPU76009.1 KPU76008.1 GEFH01002007 JAP66574.1 GEDV01007795 JAP80762.1 AXCN02000453 CM000071 EDY68850.2 CM002911 KMY94918.1 KMY94920.1 ATLV01018104 ATLV01018105 ATLV01018106 ATLV01018107 KE525212 KFB42727.1 CM000158 EDW91590.2 KRJ99976.1 KMY94921.1 KMY94919.1 KRJ99977.1 KRJ99975.1 GFXV01007327 MBW19132.1 GFDF01007140 JAV06944.1 KQ434889 KZC10232.1 GACK01009518 JAA55516.1 ABLF02031477 ABLF02031485 ACPB03001792 GFAC01004226 JAT94962.1 CH954179 KQS62101.1 EDV55121.2 CH933808 EDW09125.2 CH940648 EDW61485.2 KRF80009.1 KQS62102.1 KQS62100.1 KRG04456.1 CH963846 EDW72436.2 GDIQ01122665 JAL29061.1 LRGB01000189 KZS20507.1 KRT01765.1 GAMC01017435 JAB89120.1 OUUW01000001 SPP73500.1 KRT01763.1 KRT01764.1 JH432171 GDIP01208968 JAJ14434.1 GDIP01063740 JAM39975.1
OWR46801.1 BABH01009748 BABH01009749 BABH01009750 KQ460152 KPJ17414.1 KQ459593 KPI96325.1 DS235379 EEB15434.1 KQ414786 KOC60820.1 GBYB01012740 JAG82507.1 KQ435789 KOX74477.1 GBYB01012738 JAG82505.1 KQ981148 KYN09093.1 AE013599 BT023509 AAF57652.1 AAY84909.1 NNAY01000772 OXU26601.1 CH477261 EAT45687.1 ADTU01002507 ADTU01002508 ADTU01002509 ADTU01002510 ADTU01002511 ADTU01002512 ADTU01002513 ADTU01002514 ADTU01002515 ADTU01002516 GBHO01019479 GDHC01002131 JAG24125.1 JAQ16498.1 GL452426 EFN77247.1 KQ976747 KYM75320.1 KZ288391 PBC26364.1 GL441651 EFN64467.1 GFTR01007477 JAW08949.1 AAAB01008960 EAA11379.3 KQ982617 KYQ53809.1 KQ978350 KYM94703.1 EAU76597.1 CH480816 EDW48504.1 KK853026 KDR12311.1 KK107119 EZA58703.1 QOIP01000009 RLU18901.1 CVRI01000001 CRK86177.1 JXUM01009542 JXUM01009543 JXUM01009544 JXUM01010158 JXUM01010159 JXUM01010160 JXUM01010161 JXUM01010162 KQ560300 KQ560286 KXJ83174.1 KXJ83262.1 GACK01000070 JAA64964.1 ABV53845.2 UFQS01003371 UFQT01003371 SSX15571.1 SSX34934.1 ABV53846.2 CH916367 EDW02180.1 GECZ01000143 JAS69626.1 CH902619 EDV36111.2 GACK01009549 JAA55485.1 GGMR01005125 MBY17744.1 AHN56382.1 AAF57651.4 GBBI01003668 JAC15044.1 KPU76009.1 KPU76008.1 GEFH01002007 JAP66574.1 GEDV01007795 JAP80762.1 AXCN02000453 CM000071 EDY68850.2 CM002911 KMY94918.1 KMY94920.1 ATLV01018104 ATLV01018105 ATLV01018106 ATLV01018107 KE525212 KFB42727.1 CM000158 EDW91590.2 KRJ99976.1 KMY94921.1 KMY94919.1 KRJ99977.1 KRJ99975.1 GFXV01007327 MBW19132.1 GFDF01007140 JAV06944.1 KQ434889 KZC10232.1 GACK01009518 JAA55516.1 ABLF02031477 ABLF02031485 ACPB03001792 GFAC01004226 JAT94962.1 CH954179 KQS62101.1 EDV55121.2 CH933808 EDW09125.2 CH940648 EDW61485.2 KRF80009.1 KQS62102.1 KQS62100.1 KRG04456.1 CH963846 EDW72436.2 GDIQ01122665 JAL29061.1 LRGB01000189 KZS20507.1 KRT01765.1 GAMC01017435 JAB89120.1 OUUW01000001 SPP73500.1 KRT01763.1 KRT01764.1 JH432171 GDIP01208968 JAJ14434.1 GDIP01063740 JAM39975.1
Proteomes
UP000218220
UP000007151
UP000005204
UP000053240
UP000053268
UP000009046
+ More
UP000002358 UP000053825 UP000053105 UP000078492 UP000000803 UP000215335 UP000008820 UP000005205 UP000008237 UP000078540 UP000242457 UP000000311 UP000005203 UP000007062 UP000075809 UP000078542 UP000001292 UP000027135 UP000053097 UP000279307 UP000076408 UP000183832 UP000069940 UP000249989 UP000001070 UP000007801 UP000075886 UP000001819 UP000030765 UP000002282 UP000076502 UP000192221 UP000007819 UP000015103 UP000008711 UP000009192 UP000075920 UP000008792 UP000095300 UP000007798 UP000095301 UP000076858 UP000268350
UP000002358 UP000053825 UP000053105 UP000078492 UP000000803 UP000215335 UP000008820 UP000005205 UP000008237 UP000078540 UP000242457 UP000000311 UP000005203 UP000007062 UP000075809 UP000078542 UP000001292 UP000027135 UP000053097 UP000279307 UP000076408 UP000183832 UP000069940 UP000249989 UP000001070 UP000007801 UP000075886 UP000001819 UP000030765 UP000002282 UP000076502 UP000192221 UP000007819 UP000015103 UP000008711 UP000009192 UP000075920 UP000008792 UP000095300 UP000007798 UP000095301 UP000076858 UP000268350
Pfam
Interpro
IPR017441
Protein_kinase_ATP_BS
+ More
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR028375 KA1/Ssp2_C
IPR001772 KA1_dom
IPR000719 Prot_kinase_dom
IPR034672 SIK
IPR002999 Tudor
IPR041291 TUDOR_5
IPR016177 DNA-bd_dom_sf
IPR001739 Methyl_CpG_DNA-bd
IPR001214 SET_dom
IPR003616 Post-SET_dom
IPR041292 Tudor_4
IPR007728 Pre-SET_dom
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR028375 KA1/Ssp2_C
IPR001772 KA1_dom
IPR000719 Prot_kinase_dom
IPR034672 SIK
IPR002999 Tudor
IPR041291 TUDOR_5
IPR016177 DNA-bd_dom_sf
IPR001739 Methyl_CpG_DNA-bd
IPR001214 SET_dom
IPR003616 Post-SET_dom
IPR041292 Tudor_4
IPR007728 Pre-SET_dom
CDD
ProteinModelPortal
A0A2A4K7A3
A0A2W1BMK5
A0A2A4K5M6
A0A212EZB9
H9JPB6
A0A194RIZ9
+ More
A0A194PYU2 E0VPX8 K7ILZ4 A0A0L7QQB4 A0A0C9RV60 A0A0M9A067 A0A0C9QEA2 A0A151IQZ7 Q4QQA7 A0A232F6Z7 Q0IG70 A0A158NN80 A0A0A9Y3Q2 E2C476 A0A151HY62 A0A2A3E4E6 E2AQ24 A0A088APQ0 A0A224XLQ4 Q7Q5I9 A0A151X0P2 A0A195C3E7 A0NES6 B4HP02 A0A067R3B4 A0A026WRY8 A0A3L8DF00 A0A182YP73 A0A1J1HIR9 A0A182GE93 L7MMY1 A8DYI4 A0A336MWI8 A8DYI5 B4J7N2 A0A1B6H4M6 B3MBU0 L7LUJ6 A0A2S2NKQ2 A0A0B4LFW1 A1ZBC4 A0A023F273 A0A1S4G7H2 A0A0P8XMZ8 A0A0P8XMW6 A0A131XGE2 A0A131YQE6 A0A182QA15 B5DZ12 A0A0J9RFX1 A0A0J9U5M6 A0A084VXN3 B4P4A4 A0A0R1DS47 A0A0J9RG83 A0A0J9RHG4 A0A0R1DXK5 A0A0R1DRZ0 A0A2H8U050 A0A1L8DKS1 A0A154PEE2 L7LTW0 A0A1W4UA84 J9K8Y8 A0A1W4UNZ5 A0A1W4UA80 T1HPG7 A0A1E1X6R7 A0A0Q5VT30 B3NK99 B4KPL5 A0A1Y9IVD2 B4LJ49 A0A0Q5VKW8 A0A0Q5VLS1 A0A0Q9XKL0 A0A1I8Q3Z6 B4MJZ7 A0A0P5QAZ3 A0A1I8NCR8 A0A162RH65 A0A1I8NCR9 A0A1I8Q426 A0A0R3NS22 W8AKG4 A0A3B0JE14 A0A0R3NSH8 A0A0R3NLK6 T1JFK1 A0A0P4ZRN7 A0A0P5Y2V5
A0A194PYU2 E0VPX8 K7ILZ4 A0A0L7QQB4 A0A0C9RV60 A0A0M9A067 A0A0C9QEA2 A0A151IQZ7 Q4QQA7 A0A232F6Z7 Q0IG70 A0A158NN80 A0A0A9Y3Q2 E2C476 A0A151HY62 A0A2A3E4E6 E2AQ24 A0A088APQ0 A0A224XLQ4 Q7Q5I9 A0A151X0P2 A0A195C3E7 A0NES6 B4HP02 A0A067R3B4 A0A026WRY8 A0A3L8DF00 A0A182YP73 A0A1J1HIR9 A0A182GE93 L7MMY1 A8DYI4 A0A336MWI8 A8DYI5 B4J7N2 A0A1B6H4M6 B3MBU0 L7LUJ6 A0A2S2NKQ2 A0A0B4LFW1 A1ZBC4 A0A023F273 A0A1S4G7H2 A0A0P8XMZ8 A0A0P8XMW6 A0A131XGE2 A0A131YQE6 A0A182QA15 B5DZ12 A0A0J9RFX1 A0A0J9U5M6 A0A084VXN3 B4P4A4 A0A0R1DS47 A0A0J9RG83 A0A0J9RHG4 A0A0R1DXK5 A0A0R1DRZ0 A0A2H8U050 A0A1L8DKS1 A0A154PEE2 L7LTW0 A0A1W4UA84 J9K8Y8 A0A1W4UNZ5 A0A1W4UA80 T1HPG7 A0A1E1X6R7 A0A0Q5VT30 B3NK99 B4KPL5 A0A1Y9IVD2 B4LJ49 A0A0Q5VKW8 A0A0Q5VLS1 A0A0Q9XKL0 A0A1I8Q3Z6 B4MJZ7 A0A0P5QAZ3 A0A1I8NCR8 A0A162RH65 A0A1I8NCR9 A0A1I8Q426 A0A0R3NS22 W8AKG4 A0A3B0JE14 A0A0R3NSH8 A0A0R3NLK6 T1JFK1 A0A0P4ZRN7 A0A0P5Y2V5
PDB
2QNJ
E-value=7.79955e-100,
Score=930
Ontologies
GO
GO:0005524
GO:0004674
GO:0004683
GO:0004672
GO:0006468
GO:0005634
GO:0005737
GO:0042149
GO:0031588
GO:0032007
GO:0035556
GO:0016021
GO:0042594
GO:0042308
GO:0018107
GO:0010897
GO:0055088
GO:0008286
GO:0050802
GO:0005694
GO:0018024
GO:0003677
GO:0008270
GO:0000287
GO:0004715
GO:0007067
GO:0004713
GO:0016491
GO:0015930
Topology
Subcellular location
Chromosome
Nucleus
Nucleus
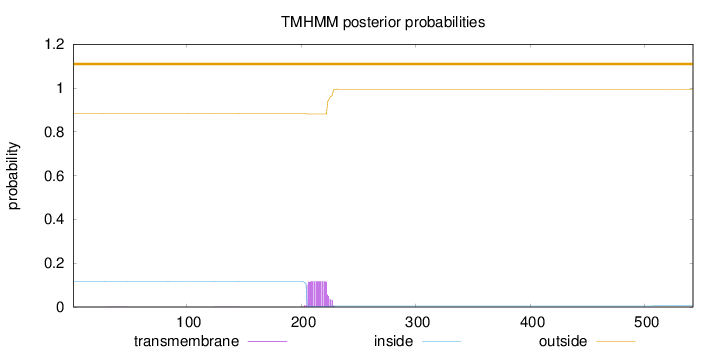
Length:
542
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.3369
Exp number, first 60 AAs:
0.0043
Total prob of N-in:
0.11591
outside
1 - 542
Population Genetic Test Statistics
Pi
188.50894
Theta
157.798856
Tajima's D
1.294746
CLR
0.250514
CSRT
0.739463026848658
Interpretation
Uncertain
