Pre Gene Modal
BGIBMGA011237
Annotation
26S_proteasome_non-ATPase_regulatory_subunit_13_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.485
Sequence
CDS
ATGGCGACTGTTAAGTTTGCAGTTATCGATGTTAGTGATTTTTTAACTAAAAAGCAAGCTAGCGAGCCTGCTTTAGCTGCGGATTGGGCGAAATTAGAGGAACTTTACAATAAAAAACTATGGCACCAACTGACTTTAAAACTGCAAGAATTCGTCAAGAATCCGGCTCTGCAGAGGGGAGACAACCTCATACAGTTGTACAATAACTTTTTGACCACCTTCGAAAGCAAAATAAATCCCCTATCATTAGTTGAGATCATTGCTCATATCGTGGAACAGTATGTCAACAAGAGAGATGCTGTCACATTCCTTGAGAAGGTGGAAACGAAAGTTAAAATGAATGATGAAGCACTGGCTCTTTGTAAGGTGCTACAAGGACAAATATATATTGAACAGCTGAATGATTATGATGCAGCTGAGAAAATAATTGAACATTTAGAAAGTACACTCGAAGATGCTGACGGTGTGACTCCAGTTCACGGACGATTCTATAAGTTAGCCTCGGAATATTACAGAGTCCGTGGTCCGACTGGCCGTTACTACCGCGCAGCGCTTCGGTACGTTGGCTGCGCCGACGGGGGTGAGGCCCTACCTGCCACGGAACGTGCTGATATCGCCTTCAGACTAGCTCTCTCTGGAGTGCTGGCTCCGGACGTATATGATTTGGGAGAATTGTTGGCTCACCCAATTCTTCATTCTCTGAGGGGCACGCCCAATGAATGGGCCTGTGAACTGGTCAAAGCTGTATCGGTTGGAGACGTTGGTGCCTTCGAGAGGATCCGGGCCAAATCCAACTGCGACGAATTACACAAAGCAGACAGGCAACTGAGACAGAAAATTGCGATTCTGTGCTTAATGGAGATGGCCTTCAATAAATCCTCATCTCAGAGGAAGCTGTCGTTCGAGGAGATAGCCCGTGAAGCCAGAATACCCCTCGACGAAGTCGAACTCCTCATCATGAAGGCTTTGGCTGAAAAGTTAATTAGAGGACATATAGATCAGGTGCGGTCGTGCGCGACGGTGCAATGGGTGCGGCCGCGTGCGCTGGCCGGGGCAGGCGCCGTGCTGACGGCAGCGCGGGTGGACGCGTGGCGAGAGGCGGTCTCCGCGGCCGCCGACCTGCTGGCTACCGTGCAGCCCGACCTCATCATCGCTTGA
Protein
MATVKFAVIDVSDFLTKKQASEPALAADWAKLEELYNKKLWHQLTLKLQEFVKNPALQRGDNLIQLYNNFLTTFESKINPLSLVEIIAHIVEQYVNKRDAVTFLEKVETKVKMNDEALALCKVLQGQIYIEQLNDYDAAEKIIEHLESTLEDADGVTPVHGRFYKLASEYYRVRGPTGRYYRAALRYVGCADGGEALPATERADIAFRLALSGVLAPDVYDLGELLAHPILHSLRGTPNEWACELVKAVSVGDVGAFERIRAKSNCDELHKADRQLRQKIAILCLMEMAFNKSSSQRKLSFEEIAREARIPLDEVELLIMKALAEKLIRGHIDQVRSCATVQWVRPRALAGAGAVLTAARVDAWREAVSAAADLLATVQPDLIIA
Summary
Uniprot
Q2F5J1
H9JNY3
A0A194PSP5
I4DQ49
S4PD39
A0A194RJ90
+ More
A0A3S2P4N4 A0A2W1BRM2 A0A2H1VPB4 A0A1Y1N2N5 E9IUG1 A0A0J7P046 A0A026WSN7 A0A0L7QWZ2 A0A195CT28 E2BGW0 A0A154PEQ4 A0A2A3EKE0 A0A151IT27 A0A158NNB4 F4WLH2 A0A151XAD5 A0A1W4WNL4 A0A0M8ZUV8 A0A195FWC4 A0A2P8XSD0 D2A3P2 A0A195AUZ7 A0A2J7QN91 A0A232FLS0 E0W1V2 R4WDY4 U4UB34 A0A1B6C6Y1 N6TBD4 A0A1B6GVK3 A0A088AD16 A0A1B6I5W3 A0A069DTT0 A0A224XE42 A0A0V0G438 A0A1B6M1C9 A0A0P4VRL8 T1HTI5 A0A1J1HJ12 R4G4S9 A0A2R7VYX1 A0A1L8DT85 A0A1L8DT72 Q16F31 A0A023ER33 A0A1S4G385 U5EVR7 A0A1B0CJ11 A0A2M3Z5S2 A0A2M4BRW4 A0A182FP76 A0A2M4AF78 W8C7E4 A0A023ES98 A0A1A9XKT5 A0A1B0AVB3 A0A0L0CBV9 W5JLM8 D3TN29 A0A1A9ZMA3 A0A034WMH5 A0A182MSQ0 A0A0K8W1I4 A0A182JYW1 A0A1Q3FE80 A0A182PGK1 B0WDK5 A0A0A1WMI5 A0A182J271 A0A182UVW5 A0A182U786 A0A182WSL4 Q7PXY1 A0A182HLG4 A0A1I8NC55 A0A0A9XLW8 A0A1A9X2C9 A0A182NS06 A0A1A9VS58 A0A182QKY1 A0A1I8QDQ4 E2AJ34 A0A0K8S4B8 A0A084VGT3 A0A182YB53 B4R239 B4HFX7 Q7KMP8 B3P8F0 B4PNW9 A0A1W4ULS6 B3LY58 A0A182RZA1
A0A3S2P4N4 A0A2W1BRM2 A0A2H1VPB4 A0A1Y1N2N5 E9IUG1 A0A0J7P046 A0A026WSN7 A0A0L7QWZ2 A0A195CT28 E2BGW0 A0A154PEQ4 A0A2A3EKE0 A0A151IT27 A0A158NNB4 F4WLH2 A0A151XAD5 A0A1W4WNL4 A0A0M8ZUV8 A0A195FWC4 A0A2P8XSD0 D2A3P2 A0A195AUZ7 A0A2J7QN91 A0A232FLS0 E0W1V2 R4WDY4 U4UB34 A0A1B6C6Y1 N6TBD4 A0A1B6GVK3 A0A088AD16 A0A1B6I5W3 A0A069DTT0 A0A224XE42 A0A0V0G438 A0A1B6M1C9 A0A0P4VRL8 T1HTI5 A0A1J1HJ12 R4G4S9 A0A2R7VYX1 A0A1L8DT85 A0A1L8DT72 Q16F31 A0A023ER33 A0A1S4G385 U5EVR7 A0A1B0CJ11 A0A2M3Z5S2 A0A2M4BRW4 A0A182FP76 A0A2M4AF78 W8C7E4 A0A023ES98 A0A1A9XKT5 A0A1B0AVB3 A0A0L0CBV9 W5JLM8 D3TN29 A0A1A9ZMA3 A0A034WMH5 A0A182MSQ0 A0A0K8W1I4 A0A182JYW1 A0A1Q3FE80 A0A182PGK1 B0WDK5 A0A0A1WMI5 A0A182J271 A0A182UVW5 A0A182U786 A0A182WSL4 Q7PXY1 A0A182HLG4 A0A1I8NC55 A0A0A9XLW8 A0A1A9X2C9 A0A182NS06 A0A1A9VS58 A0A182QKY1 A0A1I8QDQ4 E2AJ34 A0A0K8S4B8 A0A084VGT3 A0A182YB53 B4R239 B4HFX7 Q7KMP8 B3P8F0 B4PNW9 A0A1W4ULS6 B3LY58 A0A182RZA1
Pubmed
19121390
26354079
22651552
23622113
28756777
28004739
+ More
21282665 24508170 30249741 20798317 21347285 21719571 29403074 18362917 19820115 28648823 20566863 23691247 23537049 26334808 27129103 17510324 24945155 26483478 24495485 26108605 20920257 23761445 20353571 25348373 25830018 12364791 14747013 17210077 25315136 25401762 26823975 24438588 25244985 17994087 10893261 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
21282665 24508170 30249741 20798317 21347285 21719571 29403074 18362917 19820115 28648823 20566863 23691247 23537049 26334808 27129103 17510324 24945155 26483478 24495485 26108605 20920257 23761445 20353571 25348373 25830018 12364791 14747013 17210077 25315136 25401762 26823975 24438588 25244985 17994087 10893261 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
DQ311432
ABD36376.1
BABH01009736
KQ459593
KPI96332.1
AK403929
+ More
BAM20039.1 GAIX01003816 JAA88744.1 KQ460152 KPJ17405.1 RSAL01000024 RVE52235.1 KZ149962 PZC76294.1 ODYU01003649 SOQ42670.1 GEZM01014236 JAV92171.1 GL765974 EFZ15793.1 LBMM01000586 KMQ98020.1 KK107109 QOIP01000008 EZA59047.1 RLU19942.1 KQ414706 KOC63086.1 KQ977372 KYN03284.1 GL448228 EFN85092.1 KQ434889 KZC10356.1 KZ288220 PBC32160.1 KQ981037 KYN10164.1 ADTU01021202 GL888208 EGI64908.1 KQ982351 KYQ57290.1 KQ435878 KOX70009.1 KQ981208 KYN44726.1 PYGN01001429 PSN34902.1 KQ971348 EFA05541.1 KQ976738 KYM75850.1 NEVH01013198 PNF30056.1 NNAY01000050 OXU31533.1 DS235873 EEB19546.1 AK417969 BAN21184.1 KB632003 ERL87811.1 GEDC01028239 JAS09059.1 APGK01044204 KB741021 ENN75033.1 GECZ01003301 JAS66468.1 GECU01025397 JAS82309.1 GBGD01001797 JAC87092.1 GFTR01005690 JAW10736.1 GECL01003334 JAP02790.1 GEBQ01010250 JAT29727.1 GDKW01000878 JAI55717.1 ACPB03017753 CVRI01000006 CRK87907.1 GAHY01000506 JAA77004.1 KK854135 PTY11890.1 GFDF01004569 JAV09515.1 GFDF01004579 JAV09505.1 CH478517 EAT32839.1 JXUM01060246 JXUM01060247 GAPW01001988 KQ562094 JAC11610.1 KXJ76703.1 GANO01001756 JAB58115.1 AJWK01013948 GGFM01003126 MBW23877.1 GGFJ01006387 MBW55528.1 GGFK01006076 MBW39397.1 GAMC01008061 JAB98494.1 GAPW01001987 JAC11611.1 JXJN01004182 JRES01000634 KNC29740.1 ADMH02000799 ETN65026.1 CCAG010009277 EZ422831 ADD19107.1 GAKP01003098 JAC55854.1 AXCM01002120 GDHF01007350 JAI44964.1 GFDL01009182 JAV25863.1 DS231898 EDS44614.1 GBXI01014659 JAC99632.1 AAAB01008987 EAA00931.2 APCN01000897 GBHO01022993 GDHC01011201 JAG20611.1 JAQ07428.1 AXCN02000262 GL439932 EFN66566.1 GBRD01017686 JAG48141.1 ATLV01013084 KE524839 KFB37177.1 CM000364 EDX14096.1 CH480815 EDW43370.1 AF145311 AE014297 AY061232 AAF08392.1 AAF56173.1 AAF56174.1 AAL28780.1 CH954182 EDV53974.1 CM000160 EDW98179.1 CH902617 EDV43962.1
BAM20039.1 GAIX01003816 JAA88744.1 KQ460152 KPJ17405.1 RSAL01000024 RVE52235.1 KZ149962 PZC76294.1 ODYU01003649 SOQ42670.1 GEZM01014236 JAV92171.1 GL765974 EFZ15793.1 LBMM01000586 KMQ98020.1 KK107109 QOIP01000008 EZA59047.1 RLU19942.1 KQ414706 KOC63086.1 KQ977372 KYN03284.1 GL448228 EFN85092.1 KQ434889 KZC10356.1 KZ288220 PBC32160.1 KQ981037 KYN10164.1 ADTU01021202 GL888208 EGI64908.1 KQ982351 KYQ57290.1 KQ435878 KOX70009.1 KQ981208 KYN44726.1 PYGN01001429 PSN34902.1 KQ971348 EFA05541.1 KQ976738 KYM75850.1 NEVH01013198 PNF30056.1 NNAY01000050 OXU31533.1 DS235873 EEB19546.1 AK417969 BAN21184.1 KB632003 ERL87811.1 GEDC01028239 JAS09059.1 APGK01044204 KB741021 ENN75033.1 GECZ01003301 JAS66468.1 GECU01025397 JAS82309.1 GBGD01001797 JAC87092.1 GFTR01005690 JAW10736.1 GECL01003334 JAP02790.1 GEBQ01010250 JAT29727.1 GDKW01000878 JAI55717.1 ACPB03017753 CVRI01000006 CRK87907.1 GAHY01000506 JAA77004.1 KK854135 PTY11890.1 GFDF01004569 JAV09515.1 GFDF01004579 JAV09505.1 CH478517 EAT32839.1 JXUM01060246 JXUM01060247 GAPW01001988 KQ562094 JAC11610.1 KXJ76703.1 GANO01001756 JAB58115.1 AJWK01013948 GGFM01003126 MBW23877.1 GGFJ01006387 MBW55528.1 GGFK01006076 MBW39397.1 GAMC01008061 JAB98494.1 GAPW01001987 JAC11611.1 JXJN01004182 JRES01000634 KNC29740.1 ADMH02000799 ETN65026.1 CCAG010009277 EZ422831 ADD19107.1 GAKP01003098 JAC55854.1 AXCM01002120 GDHF01007350 JAI44964.1 GFDL01009182 JAV25863.1 DS231898 EDS44614.1 GBXI01014659 JAC99632.1 AAAB01008987 EAA00931.2 APCN01000897 GBHO01022993 GDHC01011201 JAG20611.1 JAQ07428.1 AXCN02000262 GL439932 EFN66566.1 GBRD01017686 JAG48141.1 ATLV01013084 KE524839 KFB37177.1 CM000364 EDX14096.1 CH480815 EDW43370.1 AF145311 AE014297 AY061232 AAF08392.1 AAF56173.1 AAF56174.1 AAL28780.1 CH954182 EDV53974.1 CM000160 EDW98179.1 CH902617 EDV43962.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000036403
UP000053097
+ More
UP000279307 UP000053825 UP000078542 UP000008237 UP000076502 UP000242457 UP000078492 UP000005205 UP000007755 UP000075809 UP000192223 UP000053105 UP000078541 UP000245037 UP000007266 UP000078540 UP000235965 UP000215335 UP000009046 UP000030742 UP000019118 UP000005203 UP000015103 UP000183832 UP000008820 UP000069940 UP000249989 UP000092461 UP000069272 UP000092443 UP000092460 UP000037069 UP000000673 UP000092444 UP000092445 UP000075883 UP000075881 UP000075885 UP000002320 UP000075880 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000095301 UP000091820 UP000075884 UP000078200 UP000075886 UP000095300 UP000000311 UP000030765 UP000076408 UP000000304 UP000001292 UP000000803 UP000008711 UP000002282 UP000192221 UP000007801 UP000075900
UP000279307 UP000053825 UP000078542 UP000008237 UP000076502 UP000242457 UP000078492 UP000005205 UP000007755 UP000075809 UP000192223 UP000053105 UP000078541 UP000245037 UP000007266 UP000078540 UP000235965 UP000215335 UP000009046 UP000030742 UP000019118 UP000005203 UP000015103 UP000183832 UP000008820 UP000069940 UP000249989 UP000092461 UP000069272 UP000092443 UP000092460 UP000037069 UP000000673 UP000092444 UP000092445 UP000075883 UP000075881 UP000075885 UP000002320 UP000075880 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000095301 UP000091820 UP000075884 UP000078200 UP000075886 UP000095300 UP000000311 UP000030765 UP000076408 UP000000304 UP000001292 UP000000803 UP000008711 UP000002282 UP000192221 UP000007801 UP000075900
Interpro
Gene 3D
ProteinModelPortal
Q2F5J1
H9JNY3
A0A194PSP5
I4DQ49
S4PD39
A0A194RJ90
+ More
A0A3S2P4N4 A0A2W1BRM2 A0A2H1VPB4 A0A1Y1N2N5 E9IUG1 A0A0J7P046 A0A026WSN7 A0A0L7QWZ2 A0A195CT28 E2BGW0 A0A154PEQ4 A0A2A3EKE0 A0A151IT27 A0A158NNB4 F4WLH2 A0A151XAD5 A0A1W4WNL4 A0A0M8ZUV8 A0A195FWC4 A0A2P8XSD0 D2A3P2 A0A195AUZ7 A0A2J7QN91 A0A232FLS0 E0W1V2 R4WDY4 U4UB34 A0A1B6C6Y1 N6TBD4 A0A1B6GVK3 A0A088AD16 A0A1B6I5W3 A0A069DTT0 A0A224XE42 A0A0V0G438 A0A1B6M1C9 A0A0P4VRL8 T1HTI5 A0A1J1HJ12 R4G4S9 A0A2R7VYX1 A0A1L8DT85 A0A1L8DT72 Q16F31 A0A023ER33 A0A1S4G385 U5EVR7 A0A1B0CJ11 A0A2M3Z5S2 A0A2M4BRW4 A0A182FP76 A0A2M4AF78 W8C7E4 A0A023ES98 A0A1A9XKT5 A0A1B0AVB3 A0A0L0CBV9 W5JLM8 D3TN29 A0A1A9ZMA3 A0A034WMH5 A0A182MSQ0 A0A0K8W1I4 A0A182JYW1 A0A1Q3FE80 A0A182PGK1 B0WDK5 A0A0A1WMI5 A0A182J271 A0A182UVW5 A0A182U786 A0A182WSL4 Q7PXY1 A0A182HLG4 A0A1I8NC55 A0A0A9XLW8 A0A1A9X2C9 A0A182NS06 A0A1A9VS58 A0A182QKY1 A0A1I8QDQ4 E2AJ34 A0A0K8S4B8 A0A084VGT3 A0A182YB53 B4R239 B4HFX7 Q7KMP8 B3P8F0 B4PNW9 A0A1W4ULS6 B3LY58 A0A182RZA1
A0A3S2P4N4 A0A2W1BRM2 A0A2H1VPB4 A0A1Y1N2N5 E9IUG1 A0A0J7P046 A0A026WSN7 A0A0L7QWZ2 A0A195CT28 E2BGW0 A0A154PEQ4 A0A2A3EKE0 A0A151IT27 A0A158NNB4 F4WLH2 A0A151XAD5 A0A1W4WNL4 A0A0M8ZUV8 A0A195FWC4 A0A2P8XSD0 D2A3P2 A0A195AUZ7 A0A2J7QN91 A0A232FLS0 E0W1V2 R4WDY4 U4UB34 A0A1B6C6Y1 N6TBD4 A0A1B6GVK3 A0A088AD16 A0A1B6I5W3 A0A069DTT0 A0A224XE42 A0A0V0G438 A0A1B6M1C9 A0A0P4VRL8 T1HTI5 A0A1J1HJ12 R4G4S9 A0A2R7VYX1 A0A1L8DT85 A0A1L8DT72 Q16F31 A0A023ER33 A0A1S4G385 U5EVR7 A0A1B0CJ11 A0A2M3Z5S2 A0A2M4BRW4 A0A182FP76 A0A2M4AF78 W8C7E4 A0A023ES98 A0A1A9XKT5 A0A1B0AVB3 A0A0L0CBV9 W5JLM8 D3TN29 A0A1A9ZMA3 A0A034WMH5 A0A182MSQ0 A0A0K8W1I4 A0A182JYW1 A0A1Q3FE80 A0A182PGK1 B0WDK5 A0A0A1WMI5 A0A182J271 A0A182UVW5 A0A182U786 A0A182WSL4 Q7PXY1 A0A182HLG4 A0A1I8NC55 A0A0A9XLW8 A0A1A9X2C9 A0A182NS06 A0A1A9VS58 A0A182QKY1 A0A1I8QDQ4 E2AJ34 A0A0K8S4B8 A0A084VGT3 A0A182YB53 B4R239 B4HFX7 Q7KMP8 B3P8F0 B4PNW9 A0A1W4ULS6 B3LY58 A0A182RZA1
PDB
6EPF
E-value=2.93768e-80,
Score=760
Ontologies
GO
PANTHER
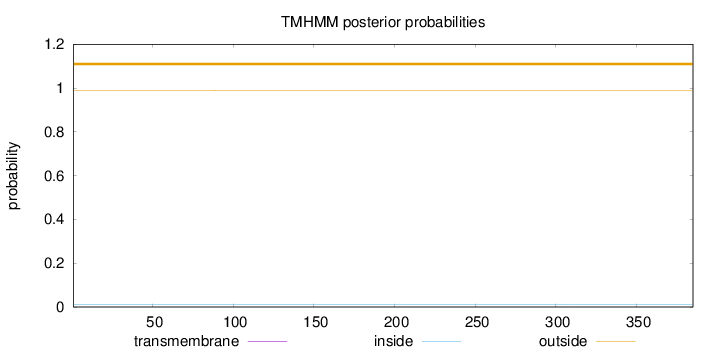
Topology
Subcellular location
Cytoplasm
Length:
385
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00371000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01159
outside
1 - 385
Population Genetic Test Statistics
Pi
277.908997
Theta
187.786615
Tajima's D
1.481743
CLR
0.444191
CSRT
0.779611019449028
Interpretation
Uncertain
