Gene
KWMTBOMO14035
Annotation
PREDICTED:_uncharacterized_protein_LOC105842300_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.904 PlasmaMembrane Reliability : 1.427
Sequence
CDS
ATGTTTTCAATAACATCTTGTACATTTTTATGTGTTTTTTTTGTACTCTGGAGTCCGGAAACATGTGCATCAGACATACCACATCTGCTCCTGTACAGCCCAGGTGATGTTCAGTACACAGACGTCCTGGTTAAGAGAGTAAATGACAATTTGACCCTCATTTGTGAGCTGCAAGGGGACAACACAAAGAGGAGATATGTCTGGAACTATGTCGCTTCGGATCTGGGAAATGTCACAGATTTTAAAACACCATACACAGTGGACACATTAGCGCCGAGCAACAGGAGTACATTGTACAAAACAGAGCTCCAGATCTCGGACAGCGGTCATTACATGTGTTCGGCCCCACCATTCAGTGTTACCAAGTATATTGTTGTACAAAATAAAGGTGCTGTGGGCTGCGCCAGGGGGGCCCTCAGCTGCGGCTTCCGTTGCGTGCTGCCGGCCTACGTGTGCGACGGCCGCACGGACTGCGCCGACGCAGAAGACGAGCACCCCCTCATGTGCGGAGAGAACGTTTGCATGCGGTCGGACCGGCTGAACTGCTCGGACGGGCGCTGCGTGCCGGCCGCGGCGTGCTGCGGGGCGCCAGCCGCGCTGTGTCGCCAGCCGGCCTGTTGCAACGAGCACCCTTCCTACTCCAGGCTCGAGGGTCCTATTGAAGTGGAGTACCCTCAAGTGTTTGATGACAGACATGCTCCAGACGACTACGGCTTCATACAATCCACCATCTACACTGTGACCGCCTGCGCCTTGATCTTCATGATAGCGGTGGTGCTGCTGATATCGGCGATATGCAAGATGCACATGAAGCGGGCCGCCCTCCGGAGCTACGCCCACGCGGAGCGCGCCACCCGCCGCCACTACAGTGTCCACTACGCACAGACCCAGCGCTTCCCGCCGTGCTACGAGGCGTCGCGTCTGCTGGAGGACGGGCGGGCCGCCTCCCCCGCGCACTCCCCGCACGACTCGCCCGCCAGACAGAACCTGCAGTGCAATCCCGGATGCCCTCCGGCCGCTGAGCTGGAGACCAGACCCGGCTTCGGGCTGGCGAGTCTGTCAGCCATCTTCTCGTCTAGATACAGACAGGTCCCACTCCCCCCCGCCGAAGTGGAGATGAACGCGATCAGGGCCTCCTCGACCAGCTCCAGCATCCAAAACTACCGGAGCCCCACGTACTGCGACCTCCAAGAGGAGCTGTTCGCGAGCCCAGAGTCTCAGGCCACGGGCAGGGACCTCAACTACATGGCCGCCCCGCTGGAACTGTTCCGCAGACGAGCGAGGAGCTCGCTAGACCGACAGCGCCTGACGTTGCAGCTGGGCCGGTTCCAGTTGACGCTACCCTTCCGGGACCGGAGACCGGACACGCCCAACGTTTCTGAAGTCGACGACGAGGACACGTACACGCTCAACGGCAGGACCATCCGGCTGCTGGGCGCCGACCTGGAGAACTACCCGGTCGTTTCAGACGGCATCGCGAGGCCGCCGCCCTACGCGGAGGCGGTTCGCTACAAGCTCTTCGGGCCGCCGCCAGAGTACCTCAGCAGGGAAGCGCTGAATCTTAGGGTCGATATGGAGGCGAGGACCAACGTGGAGATGCCGCCGCGCTACGACGATCTAGCGTCGGGCGCCTCCTCCGACAACCAACCGCGAGCCCCGGAGAGTAACGGAGTCGCCCCCACCCTGAGCTCGGTCATCGACGACCTGCCGGATATCGACAATGACGCGCCCGGCACTGCCTCCTCTCTGGCGCCGCACTAG
Protein
MFSITSCTFLCVFFVLWSPETCASDIPHLLLYSPGDVQYTDVLVKRVNDNLTLICELQGDNTKRRYVWNYVASDLGNVTDFKTPYTVDTLAPSNRSTLYKTELQISDSGHYMCSAPPFSVTKYIVVQNKGAVGCARGALSCGFRCVLPAYVCDGRTDCADAEDEHPLMCGENVCMRSDRLNCSDGRCVPAAACCGAPAALCRQPACCNEHPSYSRLEGPIEVEYPQVFDDRHAPDDYGFIQSTIYTVTACALIFMIAVVLLISAICKMHMKRAALRSYAHAERATRRHYSVHYAQTQRFPPCYEASRLLEDGRAASPAHSPHDSPARQNLQCNPGCPPAAELETRPGFGLASLSAIFSSRYRQVPLPPAEVEMNAIRASSTSSSIQNYRSPTYCDLQEELFASPESQATGRDLNYMAAPLELFRRRARSSLDRQRLTLQLGRFQLTLPFRDRRPDTPNVSEVDDEDTYTLNGRTIRLLGADLENYPVVSDGIARPPPYAEAVRYKLFGPPPEYLSREALNLRVDMEARTNVEMPPRYDDLASGASSDNQPRAPESNGVAPTLSSVIDDLPDIDNDAPGTASSLAPH
Summary
EMBL
Proteomes
Pfam
PF00057 Ldl_recept_a
Interpro
Gene 3D
CDD
ProteinModelPortal
PDB
3M0C
E-value=0.00240408,
Score=99
Ontologies
GO
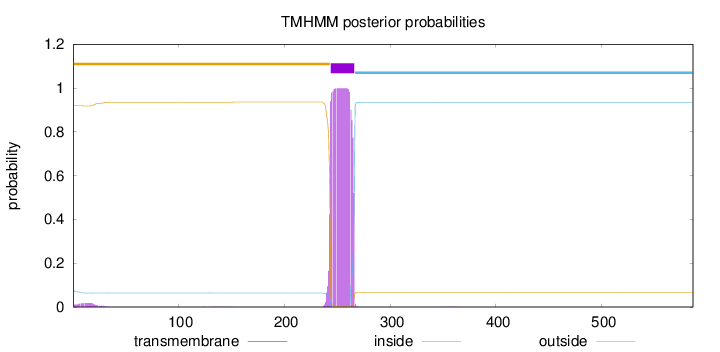
Topology
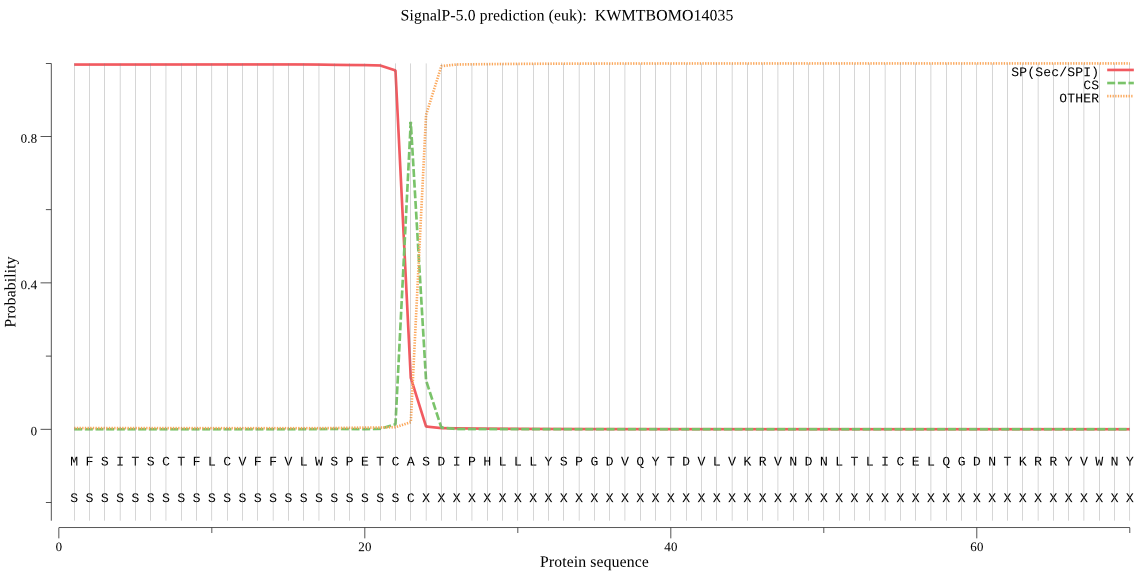
SignalP
Position: 1 - 23,
Likelihood: 0.996923
Length:
586
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.09374
Exp number, first 60 AAs:
0.36248
Total prob of N-in:
0.08074
outside
1 - 243
TMhelix
244 - 266
inside
267 - 586
Population Genetic Test Statistics
Pi
251.949379
Theta
175.903401
Tajima's D
1.171445
CLR
0.231405
CSRT
0.701464926753662
Interpretation
Uncertain
