Gene
KWMTBOMO14033
Pre Gene Modal
BGIBMGA011364
Annotation
PREDICTED:_choline/ethanolamine_kinase_isoform_X1_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 2.598
Sequence
CDS
ATGGCCAGATCGAATTCTTTCAGCATCGAAGAACCAAAGAAGGTATTGCTCAGGATATACGGCCAGGTCCACGGCGAGAGGGCCATGGACGCGATCGTGACGGAATCTGTCATATTCACTTTGCTCTCCGAGAGGAGGCTGGGCCCGAAGCTCCACGGAGTCTTCTCTGGCGGGAGGATCGAAGAGTACATACCGGCCAGACCTCTCTTGACCCGGGAACTAGCCGAGCCTGCCCTCTCTATGAAGATAGCCGAGAAGATGGCCGCCATACATTCGATGGATATTCCTCTGTCGAAGGAACCGAACTGGTTGTGGAAGACGATGTGGAAGTGGCTGAAGATCTGCAAAGACGAGAGGCTGAATACCGTCGGGAAGAACGAAGAAGAACAGCGTATCATAAAGCAATTGAAGTCGATCGATTTCAACAAAGAGATTGATTGGCTGAAGAAGTTCCTGGCTACGGTCGAGTCACCTGTCGTCTTTTGTCACAACGACATGCAAGAAGGCAACATTCTAATGCTGGAGGACTCTGCCAACGCCGACGACGAAATGACCACGATCTTTCAAGCTTATGACGTGGAGCACCACAAATCGAGCAAGGAGAACTACCACCTCGACGAGATCTCGTCCGACAGCATCTCCAGCCACATCTCGGACAGCGGCGAGCCGAAGCTCTTCCTCATCGACTTCGAGTACTGCGCTTACAACCACAGGGGCTTCGACATAGCGAACCACTTCCAGGAATGGGCCTACGACTACACGAACGACGCGCATCCGTTCTACCACGAGTGCCGGGACGACTGCCCGACCTTGGAGCAAAAGGAAATATTCATAAAGGAGTACTTAAAACATTATTCGTCGAACGTCTCCGAGGAGCCACAGGAGCCGAGCATAGATGACGTTAATCATCTGTTGAACGAGGTGGAAGCGTTCGCCCTCGCCAGCCACCTGTTCTGGACGCTCTGGTCCATTGTCAATGCTTCCAAGAGTCAGATACCCTTCGGATACTGGGAATACGCGCTGTCGAGGTTCGAAACGTACCAACGTCTCAAGATTGAGGTCATAAAGAAGGACAACTGCGGTCTGCCCCAGAAACGAAAAATATGCGAGGTCGACATATGA
Protein
MARSNSFSIEEPKKVLLRIYGQVHGERAMDAIVTESVIFTLLSERRLGPKLHGVFSGGRIEEYIPARPLLTRELAEPALSMKIAEKMAAIHSMDIPLSKEPNWLWKTMWKWLKICKDERLNTVGKNEEEQRIIKQLKSIDFNKEIDWLKKFLATVESPVVFCHNDMQEGNILMLEDSANADDEMTTIFQAYDVEHHKSSKENYHLDEISSDSISSHISDSGEPKLFLIDFEYCAYNHRGFDIANHFQEWAYDYTNDAHPFYHECRDDCPTLEQKEIFIKEYLKHYSSNVSEEPQEPSIDDVNHLLNEVEAFALASHLFWTLWSIVNASKSQIPFGYWEYALSRFETYQRLKIEVIKKDNCGLPQKRKICEVDI
Summary
Uniprot
H9JPB0
A0A3S2NMQ1
A0A2H1VVB1
A0A194PSN0
A0A194RJ09
A0A2A4JNU4
+ More
A0A2W1BRN2 A0A2J7PZV8 A0A2J7PZV9 A0A2J7PZV1 A0A3L8DIG9 A0A158NNB3 A0A0L7RDA0 A0A0J7L5X4 A0A0M8ZMP1 A0A2A3E4A6 E2AJ33 E2BGW1 F4WLH3 A0A232EZ80 A0A1S6KZC3 K7J743 A0A026WST8 D2A6I0 U5ETX9 A0A195CRB8 A0A0L0BLB3 A0A195FWN2 A0A0P4VIS2 A0A2R7X6Y1 T1HFI9 A0A151IT71 A0A067QWF6 A0A182W7I9 A0A224XKW0 A0A0V0G9H2 A0A023ESN9 A0A224XLZ2 A0A1B6EM65 T1PHW8 A0A1I8MVB7 A0A0M4E723 A0A1I8MVB5 B4LUP6 A0A0M4EHG7 A0A1I8MVB0 A0A2J7RI23 A0A023F043 A0A182M8E9 A0A0Q9X3I6 B4P6G9 A0A1D2NHB8 A0A0Q9X4L8 A0A0R1DUH1 B4N132 A0A0T6B932 A0A1W4XCD6 A0A164LLP4 A0A1W4X1K7 B4KF17 A0A0P6I2H4 A0A0P5TEN6 A0A182JSB7 B3NLR4 A0A0P6B4A1 A0A0R3NWJ1 A0A0P4VVD4 A0A0P5WQA3 A0A0Q5VYM9 A0A0P6F2T6 A0A0J9TSS8 A0A1B6CGP1 Q8IMF3 A0A1B6I5Q7 T1I4W6 A0A1I8PG92 A0A0D3QE01 A0A0D3QDH7 A0A1I8PGA3 A0A1I8PG86 A0A1I8PG18 A0A0D3QDI2 A0A0D3QDJ8 A0A0D3QE70 A0A2S2QUJ0 A0A0D3QDT5 A0A1I8PG06 A0A0D3QE66 A0A0D3QDS9 A0A0D3QE17 A0A0D3QDG6 A0A0D3QDJ4 J9KAU3 A0A0D3QDV4 A0A0K8SPA3 A0A0A9ZGX5 A0A0D3QDI7 A0A0D3QDJ0 A0A0D3QDI8 A0A0D3QDU8
A0A2W1BRN2 A0A2J7PZV8 A0A2J7PZV9 A0A2J7PZV1 A0A3L8DIG9 A0A158NNB3 A0A0L7RDA0 A0A0J7L5X4 A0A0M8ZMP1 A0A2A3E4A6 E2AJ33 E2BGW1 F4WLH3 A0A232EZ80 A0A1S6KZC3 K7J743 A0A026WST8 D2A6I0 U5ETX9 A0A195CRB8 A0A0L0BLB3 A0A195FWN2 A0A0P4VIS2 A0A2R7X6Y1 T1HFI9 A0A151IT71 A0A067QWF6 A0A182W7I9 A0A224XKW0 A0A0V0G9H2 A0A023ESN9 A0A224XLZ2 A0A1B6EM65 T1PHW8 A0A1I8MVB7 A0A0M4E723 A0A1I8MVB5 B4LUP6 A0A0M4EHG7 A0A1I8MVB0 A0A2J7RI23 A0A023F043 A0A182M8E9 A0A0Q9X3I6 B4P6G9 A0A1D2NHB8 A0A0Q9X4L8 A0A0R1DUH1 B4N132 A0A0T6B932 A0A1W4XCD6 A0A164LLP4 A0A1W4X1K7 B4KF17 A0A0P6I2H4 A0A0P5TEN6 A0A182JSB7 B3NLR4 A0A0P6B4A1 A0A0R3NWJ1 A0A0P4VVD4 A0A0P5WQA3 A0A0Q5VYM9 A0A0P6F2T6 A0A0J9TSS8 A0A1B6CGP1 Q8IMF3 A0A1B6I5Q7 T1I4W6 A0A1I8PG92 A0A0D3QE01 A0A0D3QDH7 A0A1I8PGA3 A0A1I8PG86 A0A1I8PG18 A0A0D3QDI2 A0A0D3QDJ8 A0A0D3QE70 A0A2S2QUJ0 A0A0D3QDT5 A0A1I8PG06 A0A0D3QE66 A0A0D3QDS9 A0A0D3QE17 A0A0D3QDG6 A0A0D3QDJ4 J9KAU3 A0A0D3QDV4 A0A0K8SPA3 A0A0A9ZGX5 A0A0D3QDI7 A0A0D3QDJ0 A0A0D3QDI8 A0A0D3QDU8
Pubmed
19121390
26354079
28756777
30249741
21347285
20798317
+ More
21719571 28648823 24330026 20075255 24508170 18362917 19820115 26108605 27129103 24845553 24945155 25315136 17994087 25474469 17550304 27289101 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25552603 25401762
21719571 28648823 24330026 20075255 24508170 18362917 19820115 26108605 27129103 24845553 24945155 25315136 17994087 25474469 17550304 27289101 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25552603 25401762
EMBL
BABH01009728
RSAL01000031
RVE51579.1
ODYU01004658
SOQ44775.1
KQ459593
+ More
KPI96317.1 KQ460152 KPJ17424.1 NWSH01000939 PCG73446.1 KZ149962 PZC76304.1 NEVH01020332 PNF21864.1 PNF21862.1 PNF21863.1 QOIP01000008 RLU19943.1 ADTU01021200 ADTU01021201 ADTU01021202 KQ414614 KOC68794.1 LBMM01000586 KMQ98021.1 KQ438551 KOX67207.1 KZ288427 PBC25911.1 GL439932 EFN66565.1 GL448228 EFN85093.1 GL888208 EGI64909.1 NNAY01001492 OXU23796.1 KC223173 AQT27076.1 AAZX01002970 KK107109 EZA59048.1 KQ971348 EFA05797.2 GANO01004151 JAB55720.1 KQ977372 KYN03283.1 JRES01001704 KNC20743.1 KQ981208 KYN44727.1 GDKW01002690 JAI53905.1 KK857915 PTY27548.1 ACPB03017989 KQ981037 KYN10163.1 KK852869 KDR14668.1 GFTR01005968 JAW10458.1 GECL01001306 JAP04818.1 GAPW01001293 JAC12305.1 GFTR01005588 JAW10838.1 GECZ01030775 JAS38994.1 KA647725 AFP62354.1 CP012523 ALC40246.1 CH940649 EDW63214.2 ALC40367.1 NEVH01003505 PNF40487.1 GBBI01003847 JAC14865.1 AXCM01000451 CH963920 KRF98828.1 CM000158 EDW89926.1 KRJ98792.1 LJIJ01000039 ODN04629.1 CH933807 KRG03023.1 KRJ98793.1 EDW78194.1 LJIG01009050 KRT83855.1 LRGB01003123 KZS04233.1 EDW11986.1 GDIQ01010622 JAN84115.1 GDIQ01144138 GDIP01128556 JAL07588.1 JAL75158.1 CH954179 EDV54380.1 GDIP01020620 JAM83095.1 CH379058 KRT03581.1 GDKW01000074 JAI56521.1 GDIP01095753 JAM07962.1 KQS61784.1 GDIQ01068619 JAN26118.1 CM002910 KMY91360.1 GEDC01024641 GEDC01012284 JAS12657.1 JAS25014.1 AE014134 AAN11129.1 GECU01025444 JAS82262.1 ACPB03010519 KP274783 AJC98950.1 AJC98951.1 KP274786 AJC98956.1 KP274802 AJC98988.1 AJC98989.1 GGMS01012218 MBY81421.1 AJC98957.1 KP274800 AJC98984.1 KP274784 AJC98952.1 AJC98985.1 AJC98953.1 KP274799 AJC98983.1 ABLF02031350 ABLF02031359 AJC98982.1 GBRD01011169 JAG54655.1 GBHO01002269 JAG41335.1 KP274788 AJC98961.1 KP274797 AJC98978.1 KP274791 AJC98966.1 KP274794 AJC98972.1
KPI96317.1 KQ460152 KPJ17424.1 NWSH01000939 PCG73446.1 KZ149962 PZC76304.1 NEVH01020332 PNF21864.1 PNF21862.1 PNF21863.1 QOIP01000008 RLU19943.1 ADTU01021200 ADTU01021201 ADTU01021202 KQ414614 KOC68794.1 LBMM01000586 KMQ98021.1 KQ438551 KOX67207.1 KZ288427 PBC25911.1 GL439932 EFN66565.1 GL448228 EFN85093.1 GL888208 EGI64909.1 NNAY01001492 OXU23796.1 KC223173 AQT27076.1 AAZX01002970 KK107109 EZA59048.1 KQ971348 EFA05797.2 GANO01004151 JAB55720.1 KQ977372 KYN03283.1 JRES01001704 KNC20743.1 KQ981208 KYN44727.1 GDKW01002690 JAI53905.1 KK857915 PTY27548.1 ACPB03017989 KQ981037 KYN10163.1 KK852869 KDR14668.1 GFTR01005968 JAW10458.1 GECL01001306 JAP04818.1 GAPW01001293 JAC12305.1 GFTR01005588 JAW10838.1 GECZ01030775 JAS38994.1 KA647725 AFP62354.1 CP012523 ALC40246.1 CH940649 EDW63214.2 ALC40367.1 NEVH01003505 PNF40487.1 GBBI01003847 JAC14865.1 AXCM01000451 CH963920 KRF98828.1 CM000158 EDW89926.1 KRJ98792.1 LJIJ01000039 ODN04629.1 CH933807 KRG03023.1 KRJ98793.1 EDW78194.1 LJIG01009050 KRT83855.1 LRGB01003123 KZS04233.1 EDW11986.1 GDIQ01010622 JAN84115.1 GDIQ01144138 GDIP01128556 JAL07588.1 JAL75158.1 CH954179 EDV54380.1 GDIP01020620 JAM83095.1 CH379058 KRT03581.1 GDKW01000074 JAI56521.1 GDIP01095753 JAM07962.1 KQS61784.1 GDIQ01068619 JAN26118.1 CM002910 KMY91360.1 GEDC01024641 GEDC01012284 JAS12657.1 JAS25014.1 AE014134 AAN11129.1 GECU01025444 JAS82262.1 ACPB03010519 KP274783 AJC98950.1 AJC98951.1 KP274786 AJC98956.1 KP274802 AJC98988.1 AJC98989.1 GGMS01012218 MBY81421.1 AJC98957.1 KP274800 AJC98984.1 KP274784 AJC98952.1 AJC98985.1 AJC98953.1 KP274799 AJC98983.1 ABLF02031350 ABLF02031359 AJC98982.1 GBRD01011169 JAG54655.1 GBHO01002269 JAG41335.1 KP274788 AJC98961.1 KP274797 AJC98978.1 KP274791 AJC98966.1 KP274794 AJC98972.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000218220
UP000235965
+ More
UP000279307 UP000005205 UP000053825 UP000036403 UP000053105 UP000242457 UP000000311 UP000008237 UP000007755 UP000215335 UP000002358 UP000053097 UP000007266 UP000078542 UP000037069 UP000078541 UP000015103 UP000078492 UP000027135 UP000075920 UP000095301 UP000092553 UP000008792 UP000075883 UP000007798 UP000002282 UP000094527 UP000009192 UP000192223 UP000076858 UP000075881 UP000008711 UP000001819 UP000000803 UP000095300 UP000007819
UP000279307 UP000005205 UP000053825 UP000036403 UP000053105 UP000242457 UP000000311 UP000008237 UP000007755 UP000215335 UP000002358 UP000053097 UP000007266 UP000078542 UP000037069 UP000078541 UP000015103 UP000078492 UP000027135 UP000075920 UP000095301 UP000092553 UP000008792 UP000075883 UP000007798 UP000002282 UP000094527 UP000009192 UP000192223 UP000076858 UP000075881 UP000008711 UP000001819 UP000000803 UP000095300 UP000007819
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JPB0
A0A3S2NMQ1
A0A2H1VVB1
A0A194PSN0
A0A194RJ09
A0A2A4JNU4
+ More
A0A2W1BRN2 A0A2J7PZV8 A0A2J7PZV9 A0A2J7PZV1 A0A3L8DIG9 A0A158NNB3 A0A0L7RDA0 A0A0J7L5X4 A0A0M8ZMP1 A0A2A3E4A6 E2AJ33 E2BGW1 F4WLH3 A0A232EZ80 A0A1S6KZC3 K7J743 A0A026WST8 D2A6I0 U5ETX9 A0A195CRB8 A0A0L0BLB3 A0A195FWN2 A0A0P4VIS2 A0A2R7X6Y1 T1HFI9 A0A151IT71 A0A067QWF6 A0A182W7I9 A0A224XKW0 A0A0V0G9H2 A0A023ESN9 A0A224XLZ2 A0A1B6EM65 T1PHW8 A0A1I8MVB7 A0A0M4E723 A0A1I8MVB5 B4LUP6 A0A0M4EHG7 A0A1I8MVB0 A0A2J7RI23 A0A023F043 A0A182M8E9 A0A0Q9X3I6 B4P6G9 A0A1D2NHB8 A0A0Q9X4L8 A0A0R1DUH1 B4N132 A0A0T6B932 A0A1W4XCD6 A0A164LLP4 A0A1W4X1K7 B4KF17 A0A0P6I2H4 A0A0P5TEN6 A0A182JSB7 B3NLR4 A0A0P6B4A1 A0A0R3NWJ1 A0A0P4VVD4 A0A0P5WQA3 A0A0Q5VYM9 A0A0P6F2T6 A0A0J9TSS8 A0A1B6CGP1 Q8IMF3 A0A1B6I5Q7 T1I4W6 A0A1I8PG92 A0A0D3QE01 A0A0D3QDH7 A0A1I8PGA3 A0A1I8PG86 A0A1I8PG18 A0A0D3QDI2 A0A0D3QDJ8 A0A0D3QE70 A0A2S2QUJ0 A0A0D3QDT5 A0A1I8PG06 A0A0D3QE66 A0A0D3QDS9 A0A0D3QE17 A0A0D3QDG6 A0A0D3QDJ4 J9KAU3 A0A0D3QDV4 A0A0K8SPA3 A0A0A9ZGX5 A0A0D3QDI7 A0A0D3QDJ0 A0A0D3QDI8 A0A0D3QDU8
A0A2W1BRN2 A0A2J7PZV8 A0A2J7PZV9 A0A2J7PZV1 A0A3L8DIG9 A0A158NNB3 A0A0L7RDA0 A0A0J7L5X4 A0A0M8ZMP1 A0A2A3E4A6 E2AJ33 E2BGW1 F4WLH3 A0A232EZ80 A0A1S6KZC3 K7J743 A0A026WST8 D2A6I0 U5ETX9 A0A195CRB8 A0A0L0BLB3 A0A195FWN2 A0A0P4VIS2 A0A2R7X6Y1 T1HFI9 A0A151IT71 A0A067QWF6 A0A182W7I9 A0A224XKW0 A0A0V0G9H2 A0A023ESN9 A0A224XLZ2 A0A1B6EM65 T1PHW8 A0A1I8MVB7 A0A0M4E723 A0A1I8MVB5 B4LUP6 A0A0M4EHG7 A0A1I8MVB0 A0A2J7RI23 A0A023F043 A0A182M8E9 A0A0Q9X3I6 B4P6G9 A0A1D2NHB8 A0A0Q9X4L8 A0A0R1DUH1 B4N132 A0A0T6B932 A0A1W4XCD6 A0A164LLP4 A0A1W4X1K7 B4KF17 A0A0P6I2H4 A0A0P5TEN6 A0A182JSB7 B3NLR4 A0A0P6B4A1 A0A0R3NWJ1 A0A0P4VVD4 A0A0P5WQA3 A0A0Q5VYM9 A0A0P6F2T6 A0A0J9TSS8 A0A1B6CGP1 Q8IMF3 A0A1B6I5Q7 T1I4W6 A0A1I8PG92 A0A0D3QE01 A0A0D3QDH7 A0A1I8PGA3 A0A1I8PG86 A0A1I8PG18 A0A0D3QDI2 A0A0D3QDJ8 A0A0D3QE70 A0A2S2QUJ0 A0A0D3QDT5 A0A1I8PG06 A0A0D3QE66 A0A0D3QDS9 A0A0D3QE17 A0A0D3QDG6 A0A0D3QDJ4 J9KAU3 A0A0D3QDV4 A0A0K8SPA3 A0A0A9ZGX5 A0A0D3QDI7 A0A0D3QDJ0 A0A0D3QDI8 A0A0D3QDU8
PDB
3LQ3
E-value=6.0171e-72,
Score=688
Ontologies
PATHWAY
GO
PANTHER
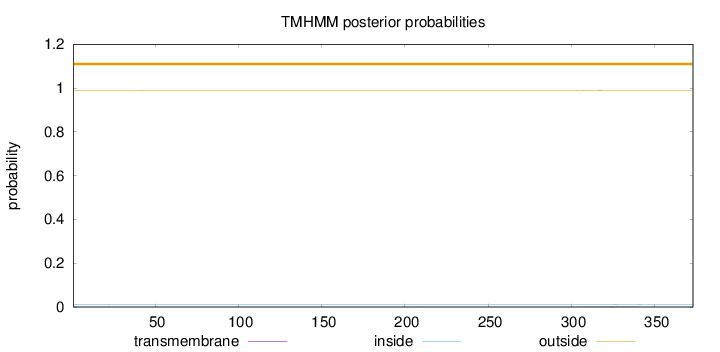
Topology
Length:
373
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04657
Exp number, first 60 AAs:
0.00195
Total prob of N-in:
0.01068
outside
1 - 373
Population Genetic Test Statistics
Pi
167.877585
Theta
159.305251
Tajima's D
0.037054
CLR
153.067196
CSRT
0.378681065946703
Interpretation
Uncertain
