Gene
KWMTBOMO14032 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011362
Annotation
hypothetical_protein_KGM_02677_[Danaus_plexippus]
Location in the cell
Extracellular Reliability : 1.537 Nuclear Reliability : 1.459
Sequence
CDS
ATGAAGCCACTATTTTATATAGTGGTGACCTGCTTGATACTGGCAATCAAAGCCCACCCCGAGAACATTGAAGATGTCAAGGATCTGCCCCAAGCCAATGAGCCAGTACTTGAGGCCGAAGAAAGCGAACCTCAAGCGAGGATAGAACGTTGCACCACATGTACGCAAAACCTCAAGCTCAGCTCCCCTACAGACGTTCTGGCTGCCATCAAGAACCTGCCTGGTGAAGTCCACACACAGCAGTCTTTCGAAGGCTGCTCCACAGAGAAAGGCTGCGCTGGTATTAAGCTGAAGGACGGAAAAGTCGTCGAGAAATTCGGGGACGTTGAGTCGTTCAAGAATGCGGCCGCTCTAGACAAATCCAAGGAATTCACTTTCCACCAAGCTGGAACGTTCGGGAAACTCATCGAGGGAAAAATACCCGACAACGGTCCCTTCTGGTGGATGAATCAAAACTCCCCCTTCAAGAACACTGGAGGCTTCGAGAAGTTCAGCAAAGCGAGTTCGAGCTACACCAGCGCCACGGGTGGTGGAATCGATCTGGCCGGGAACCCGTTCCTGAACGGTGACTTCTCCAAGTTGGGTGCTGGCTTCGCTGGCGCCGGTGCTGTTGGACAACCGACCAACAACTTCCAGTCTTCGGCATTTGAGTCTTCCTCCACTTACAGCAGCTCAGGGGGACTTGATGTGTCCGGCAACCCGTTTTTAAATGGAGGAGTTAGGTTGGGTCAAGGAGGAATCGGAGGTGGGCAAGGCGTCGCCGGCGCTCAGGGCATCGGCGGCGGCCAAATTGGCCAAGGTTTTGCCCAAAACGCCTTTGCCGGCAGCGCTCAAAATGGCTTCGGCTACACTGGGTCCTCGCCCAGACCGTTCTCCGCTTCCACGCCCGGGTCCAACGTGAACCTCATCCAGAACGCGCAGAAGGGCGGTGGATACGAATACGAACAGAACGCCCAGAGCGTCGACGAAGCATTCCAGAGTACTGGGAACGTTATAACTCCGGATAATGGAGGGGAGATACAGCAGACCTGCGCCGGGCAGGGCTACGTGTGCGTCCCGCGCGCCCAGTGCAACAACGGCATTGTCAGTGCCAACGGAGCGGCGTTACTCCAAGCCAACACGCAGAAACAATACTGCAACGTACGAAGCGAAATCTGCTGCAGAATCGAGATCTCCGGTCTTGCTGGAGCTGGCTCTATCCAAGGCTCTCTCGGAGTAGACTCTGCTTCCTACTCTGGTCAGGGAACGAATCTTTTCGCAACGAATCAAGGAGCTGCGAACAACTTTGGAGCCTTCGATTCCGCAAACAGAGCTTCTAGTTTTGGAGCGTCCAACTTCGGAAATGGTGCTGCAGTGAACAGAGCTTTTGATTCAAGAGGAGCATCAGGCATCTCTACTATTGGGTCGACTCTCGTTCCCCCCACTACAGCCGGTCGCTTCGGCTCTACTGGTTTTGGCAACGAAGTCTTCAAAGCGACAAGCCAATCGAACTTTATCGAAACCGATTCATTCTCCGCCGGTAGTGACGTGGCCAGTTCTTACAGACCAGGCGCAATCGGCTCCGGATTGAAACCCGGTATTCCCTATCTACCACCAGTCGACAACACCAACAGTGGATCTAATGTGGTGACCAGCACGGCTTTCCCGACGACCTTCGTGACGACGCCGCGTCCGTTCTCAACTCCCCGTCCGGTTACTCCTAAGCCCACTTACCTACCGCCCATCGTACCTTCGACATCTGCCCCTGGATACTTGCCTCCCGTAGCAGACTCATCCAACAACAGGGAGACCCGTCTTCCCCCTAACAAGGTTTACAATGAGGGAAGCGTGATCTTGGACGAAACAAGACAGCCTACGACCAGACCAACAGTTTTCACTCCAGGTCCTTCACCAAGTGAAATTCCTGCTGGATGCGCTGCTGCCCTCAAGTGTACTCCTATCGAATTCTGCACAGCTGAAGGTGTCATCTCAAACACTTCAGTCATCCTGAGTCGCGAACAAGACGCGTACCGAGTACCCTTGACAGACTGCAAAGACCTGGGGTCGGGTCGTATCGGGAAATGCTGCCGCGATCCCTTCTACACTGATCCATGGCCAACGAACCAGCTGGGCAAGTGGGTTCCCGGTGTCTTCGGAGGCAACGACGGCAAATACGTACCAGACAGTCGCGTCAGCCCGTCTACTAACGTGCGACCTCAAGTCACCGGCAGACCCCCCACCTCTCCTTCCACTGGATCAGTCATCTTGAACAACTTCAGGAATACCCCTAAACCAATTTCACCAACACCTGGACCCAACCAACTGTCTCCTGGTTACCCGTCGAGCACCCTGGCTCCTGCCTTCGTGCAGAAGAATCAGTTCAACCAAGGATCGTACGGTCAGAAGGGCGTTGGGCAAGGCAGCGTTATCGGAGTTGGACAAAACGGCCAAGGTAACCAAGGCTCTTATGGCCAAGGTTCGCAATTCACGCAAACCTCATCATCGAGAGGCCAAGGACAGTTCCCAGGTGTTGGCCAAGGACAGTTCCCAGGTGCTGGCCAAGGGCAGTTCCCAGGTGCTGGCCAAGGACAGTTCCCAGGTGCTGGCCAAGGACAGTTCCCAGGTGCTGGCCAAGGACAGTTCCCAGGTGCTGGCCAAGGGCAGTTCCCAGGTGCTGGCCAAGGGCAGTTCCCAGGTGCTGGCCAAGGAAAGTTCCCAGGTGCTGGCCAAGGCCAGTTTCCAGGTGCTGGCCAAACCGGGATACAAGGAGTTGATGTTGGCGTCACTCAAGGATTTGGCCAAGGTGCCTCCCAAGGAATTGGCCAACAAGCCATTTTCGGTCAAGGAAACAGAGGCCAATTTGGCCAAGGCACCCAGACTGTCTTCGGACAAGGACAGGGGGTCAGCCAAAAAAAAGGTTTTGGAGTCTCTCAAGGCGCTGGAGTCGGTATCAGGAACCAAGGAGCCGGCCAAGGTACACGTGTTACCTCTGACCAAGGCCAAGTATACGACCAAGGTGTTGTCCAAACCGTACAACAGGGCGTCGTCACCGCAGCGCAGCAGGGCGCAGGACAAGGAGTTAAGTACGGCCAAGGTCAAGGCTTCCAACAAGGAGGTGGCGTTAGCGTCAGTCAAGGAAGCTATGGTCAAGGAGAGTCACAGGGTGCCGGCATCAGTGCAAGCCGTGGCTCCGGACTGGCTGTCAATAAAGGAGCTGGCTTCGGAGTTAACCAAGGCACCGGTCAAGGCCAAAGTGGAGGATTCGGTGTTAGTCAGTCTACCGGTCAAGGTTTCAGTCAAGGCACTGGTTTAGGTGTCAACCAAGGAACGGAAGAAGGAATTATCCAAAATGGCGGTTTCGGGGTTGTCAGAGGATCTGGAACTGCCATCACCCAAGGAGGTGGATTTGCAATCAGCCAAGAAACAGGTCAAGAAAGCAGCTTCAGGGTTAACCAAGGTAGCGCTGGACAAGGAATATCTCAGAGCGGTGGCTTCGGAAACGGCCAAGGTGCGGGTCAAGGAGGCTTCGGGCAAGGTGGCAGCTTCGGTGTTGTTCAAGGAGGCGGATTCGGAGTAAGAGGTCAAGGCTTTGGCGTGAACCGCGGTGTAGGCATCGCGGTCACAGGTGGTCAAGGAGAAGCGATAAATCAAGGCTTTGGCCAAGGTGTTTACAACGGCGGTGGTTTCGGTACGGGCCAGCGTCTGGTCTCCGGCCGAGGGCAAGCGCTCGTCCAGGGCGAAGGTCAGACAGTCATCCAGGGAGTCGGCACCGGGGTCAGACAAAGCCAGGGTTTCACCGTCACACAGGGCAACGGCGTTGGAGTCGAGAATGAGTACAGTGCCTCAACGCAGCGGGTGTTCTTGAAGAGATACTCCGGCGGTGGACAATGTGGGCTTTTGAATCCCCAGAAACCGTACGGCAACCGAAATGATCTAGAAGTAGACTTCGCGGAGATACCCTGGCAAGCGATGGTGCTTCTCCAGACCAACAGGAGCCTGCTGTGCGGAGGAGTCATCACCAGGCCTGAGGTGGTTGTCACTTCAGCATCTTGCGTCGATGGGCTGGACGCGAAAAACGTTCTGATCAAAGGCGGAGAATGGAAACTCGGGATTGACGACGAGCCCCTCCCCTTCCAAATAGTCCAGGTCAAGACCATCATCCGCCACCCGGAGTACAAGAGGGGAAGCCTGAAATACGACGCAGCCATACTGGTGCTCTCGGAGAATCTGCGGTTCGCGAAGAACATCCAACCGATATGTCTCCCCGCAGCCGGAGAGACCTTGGACGCGTATTACAATGGAGCTGGAGAATGCATGGTCACTGGCTGGGGAAAGATCGTCTTGCAAGCTCATTTGTCCGGAAGCATCATGCATTCCCTAAACGTTTCTCTCATCAATCCTGGTGAGTGCGAAGCCAAGCTGTCCCAGGATTACCCCCACCTGCTGGAGCAGTACGACCAGGATAGCTGTGCATGCGGGCAGCCCACCAACCCACTCAACAATATCTGCAAGGTTGACATTGGCAGTGCCCTGGCGTGCACAACAGGAGACAGCCATTTCGTGCTACGAGGAGTCTACTCTTGGGACTCTGGTTGTCAGGTCGGTAACCAAATAGCGGGCTTCTACAAGTTCGACATTGAGTGGTACGAATGGGCCATAGGACTGATCGAGAGCGTTCGTTTCACTAAATACACGATCGGTACGAAGTTGACGCAGAGCAAGATCACCAGTCAAGTTTCCAGCGTCAAAACCAGTCAGTACACAGCCGGAGTTAAGGGCGTTAATGAAGTTGGTCAGGTTAAAGCTGACGGTTTGGCTCAGGGACAATTCGCTGTGAAAGACGACGGGTTCGCTCAAAGGCAATTCGCCGCTAAAGACGACAAGTTCGCTCTAGGGCAATTCGCTGCCAAAGGCGACGGGTTCGCTCAAGGACAATTCGCTGCCAAAGACGACAGATTCGCTCAAGGGCAATTCGCCGCGAAAACCGACTCGAAAACCCAAGGTTTCACCGGACAATTCAACCAGTACGGAACTAAATTCTCCGAGGTGAAAGCCGGCGAGGGCATAAGCTTCGACACCCAAGTCAAGGGTCCCATATCAAACACTTTCAGCGCGACCTACACCGAGAAGAAGGTGTACCAGTCCGAGCCGAAGATAGTGACGTACACGACGAAACCAGAGATCGTGACCTTCACCACGAAACCGGAGTACTTCACTTACACGACCAAACCGAAAATCGTGCGCTACACGACCAAACCAGAGATCGTGACCTTCACAACGAAACCCGAATACTTCACGTACACGACCAAACCGAAAATCGTTACCTACACCACGAAGCCGCAACTCATAACGTACGAGACTTCTGGAAGTGGAACGAACCCCCAATACGTGGCCCCGGGGGTGTCCTTCAACCCGTCCTTGACAGACGCCCTGCACAAACACGACGGCCAATGCAAATGCTTGGAGGGTAAAAAATAA
Protein
MKPLFYIVVTCLILAIKAHPENIEDVKDLPQANEPVLEAEESEPQARIERCTTCTQNLKLSSPTDVLAAIKNLPGEVHTQQSFEGCSTEKGCAGIKLKDGKVVEKFGDVESFKNAAALDKSKEFTFHQAGTFGKLIEGKIPDNGPFWWMNQNSPFKNTGGFEKFSKASSSYTSATGGGIDLAGNPFLNGDFSKLGAGFAGAGAVGQPTNNFQSSAFESSSTYSSSGGLDVSGNPFLNGGVRLGQGGIGGGQGVAGAQGIGGGQIGQGFAQNAFAGSAQNGFGYTGSSPRPFSASTPGSNVNLIQNAQKGGGYEYEQNAQSVDEAFQSTGNVITPDNGGEIQQTCAGQGYVCVPRAQCNNGIVSANGAALLQANTQKQYCNVRSEICCRIEISGLAGAGSIQGSLGVDSASYSGQGTNLFATNQGAANNFGAFDSANRASSFGASNFGNGAAVNRAFDSRGASGISTIGSTLVPPTTAGRFGSTGFGNEVFKATSQSNFIETDSFSAGSDVASSYRPGAIGSGLKPGIPYLPPVDNTNSGSNVVTSTAFPTTFVTTPRPFSTPRPVTPKPTYLPPIVPSTSAPGYLPPVADSSNNRETRLPPNKVYNEGSVILDETRQPTTRPTVFTPGPSPSEIPAGCAAALKCTPIEFCTAEGVISNTSVILSREQDAYRVPLTDCKDLGSGRIGKCCRDPFYTDPWPTNQLGKWVPGVFGGNDGKYVPDSRVSPSTNVRPQVTGRPPTSPSTGSVILNNFRNTPKPISPTPGPNQLSPGYPSSTLAPAFVQKNQFNQGSYGQKGVGQGSVIGVGQNGQGNQGSYGQGSQFTQTSSSRGQGQFPGVGQGQFPGAGQGQFPGAGQGQFPGAGQGQFPGAGQGQFPGAGQGQFPGAGQGQFPGAGQGKFPGAGQGQFPGAGQTGIQGVDVGVTQGFGQGASQGIGQQAIFGQGNRGQFGQGTQTVFGQGQGVSQKKGFGVSQGAGVGIRNQGAGQGTRVTSDQGQVYDQGVVQTVQQGVVTAAQQGAGQGVKYGQGQGFQQGGGVSVSQGSYGQGESQGAGISASRGSGLAVNKGAGFGVNQGTGQGQSGGFGVSQSTGQGFSQGTGLGVNQGTEEGIIQNGGFGVVRGSGTAITQGGGFAISQETGQESSFRVNQGSAGQGISQSGGFGNGQGAGQGGFGQGGSFGVVQGGGFGVRGQGFGVNRGVGIAVTGGQGEAINQGFGQGVYNGGGFGTGQRLVSGRGQALVQGEGQTVIQGVGTGVRQSQGFTVTQGNGVGVENEYSASTQRVFLKRYSGGGQCGLLNPQKPYGNRNDLEVDFAEIPWQAMVLLQTNRSLLCGGVITRPEVVVTSASCVDGLDAKNVLIKGGEWKLGIDDEPLPFQIVQVKTIIRHPEYKRGSLKYDAAILVLSENLRFAKNIQPICLPAAGETLDAYYNGAGECMVTGWGKIVLQAHLSGSIMHSLNVSLINPGECEAKLSQDYPHLLEQYDQDSCACGQPTNPLNNICKVDIGSALACTTGDSHFVLRGVYSWDSGCQVGNQIAGFYKFDIEWYEWAIGLIESVRFTKYTIGTKLTQSKITSQVSSVKTSQYTAGVKGVNEVGQVKADGLAQGQFAVKDDGFAQRQFAAKDDKFALGQFAAKGDGFAQGQFAAKDDRFAQGQFAAKTDSKTQGFTGQFNQYGTKFSEVKAGEGISFDTQVKGPISNTFSATYTEKKVYQSEPKIVTYTTKPEIVTFTTKPEYFTYTTKPKIVRYTTKPEIVTFTTKPEYFTYTTKPKIVTYTTKPQLITYETSGSGTNPQYVAPGVSFNPSLTDALHKHDGQCKCLEGKK
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
Topology
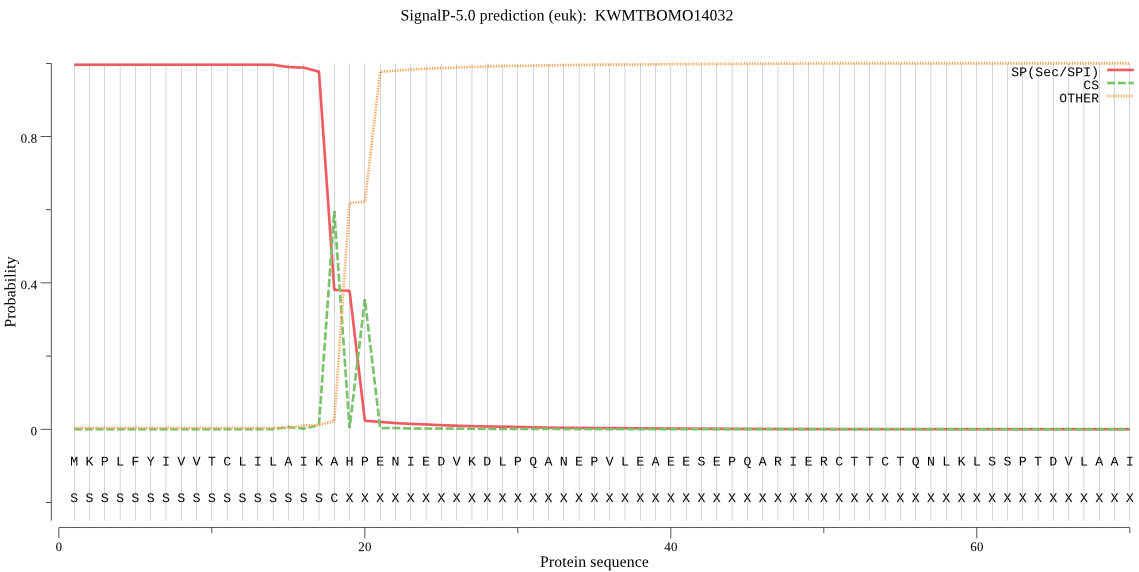
SignalP
Position: 1 - 18,
Likelihood: 0.995906
Length:
1818
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03014
Exp number, first 60 AAs:
0.02962
Total prob of N-in:
0.00169
outside
1 - 1818

Population Genetic Test Statistics
Pi
171.957053
Theta
146.079047
Tajima's D
0.502848
CLR
1.012114
CSRT
0.510724463776811
Interpretation
Uncertain
