Pre Gene Modal
BGIBMGA011360
Annotation
Putative_glycogen_[starch]_synthase_[Papilio_machaon]
Full name
Glycogen [starch] synthase
Alternative Name
Glycogen synthase
Location in the cell
Cytoplasmic Reliability : 2.017
Sequence
CDS
ATGTCAAAGGATCGCAGCTCGAGGCGGTTCTATCGTGCGGAGTCCACTCATGATCTGTCGTCATTCATGGACCGCGGCCGCGCCGCCAGCGACGAGAACAGATGGACCTTCGAGTGCGCCTGGGAAGTCGCCAACAAAGTTGGCGGTATTTATACGGTAATAAGATCGAAAGCATTCGTCTCCACTGAAGAAATGGGAGACAACTATTGCTTGCTTGGTCCGTATAAGGAGCAATGCGCGCGAACTGAAGTCGAAGAGGGAGAGTTCCCCTCCGACTCCCCCCTGTTGACAGCCGTTAACGCCATGAGGAGTAAAGGTTACAAGTTACACACGGGCACTTGGCTGGTAGATGGGAACCCTCAAATAGTTCTTTTCGACATCGGGTCTGGGGCGTGGCAGTTGGATGCTTACAAGCAGGAGCTATGGGACACTTGCTCCGTCGGCATCCCGCATCTCGACGTGGAGGCCAACGACGCCGTGATCCTAGGGTTCATGGTCGCGCAGTTCATAAGCGAGTTCCGCGAGGCGGTGGAGGGGCGCGGTGGGGTGCCGGCTCGCGTGGTGGCGCAGTTCCACGAGTGGCAGGCGGGAATCGGGCTCATCGTTCTAAGGGTACGCCATGTTGACGTGGCGACCGTATTCACGACTCACGCGACGCTCCTCGGCAGATACCTCTGCGCTGGCAACACAGACTTCTACAATAACCTGGATAAGTTCTCTGTGGACGAGGAGGCGGGAAAGAGACAGATATACCACCGCTACTGCATGGAGCGGGCCGCCTCGCACATGGCACACATCTTCACCACCGTCTCCGACATCACGGGATACGAAGCCGAGTACTTGCTCAAACGGAAGCCGGACATCATCACCCCGAACGGTTTGAACGTGAAGAAATTCTCGGCGCTGCACGAGTTCCAGAACCTGCACGCCCTCGCCAAGGACAAGATACACGAATTCGTTAGGGGACATTTTTACGGGTAA
Protein
MSKDRSSRRFYRAESTHDLSSFMDRGRAASDENRWTFECAWEVANKVGGIYTVIRSKAFVSTEEMGDNYCLLGPYKEQCARTEVEEGEFPSDSPLLTAVNAMRSKGYKLHTGTWLVDGNPQIVLFDIGSGAWQLDAYKQELWDTCSVGIPHLDVEANDAVILGFMVAQFISEFREAVEGRGGVPARVVAQFHEWQAGIGLIVLRVRHVDVATVFTTHATLLGRYLCAGNTDFYNNLDKFSVDEEAGKRQIYHRYCMERAASHMAHIFTTVSDITGYEAEYLLKRKPDIITPNGLNVKKFSALHEFQNLHALAKDKIHEFVRGHFYG
Summary
Description
Transfers the glycosyl residue from UDP-Glc to the non-reducing end of alpha-1,4-glucan.
Transfers the glycosyl residue from UDPG to the non-reducing end of alpha-1,4-glucan. In larval skeletal muscle, isoform B is required for the formation of autophagosomes during starvation and during cloroquine-induced vacuolar myopathy.
Transfers the glycosyl residue from UDPG to the non-reducing end of alpha-1,4-glucan. In larval skeletal muscle, isoform B is required for the formation of autophagosomes during starvation and during cloroquine-induced vacuolar myopathy.
Catalytic Activity
[(1->4)-alpha-D-glucosyl](n) + UDP-alpha-D-glucose = [(1->4)-alpha-D-glucosyl](n+1) + H(+) + UDP
Subunit
Isoform B interacts with Atg8a upon starvation.
Similarity
Belongs to the glycosyltransferase 3 family.
Keywords
Alternative splicing
Complete proteome
Cytoplasmic vesicle
Glycogen biosynthesis
Glycosyltransferase
Phosphoprotein
Reference proteome
Transferase
Feature
chain Glycogen [starch] synthase
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JPA6
A0A194RQV1
A0A2A4IX43
A0A2H1VWK1
A0A212EVT8
F2Q6J8
+ More
A0A154P9X4 A0A195C3G8 A0A195AVG9 A0A026X010 A0A195ETM8 A0A3L8E0G6 A0A2J7RI90 A0A151WGI9 A0A195E342 A0A2J7RI94 F4WA55 A0A0N0BF19 A0A067QZJ4 E2BGV0 A0A1B6CWL5 C0KJJ5 A0A088ARU9 E2A8G5 A0A1B6KQ54 A0A1B6GZ27 A0A0L7RDK2 E0VIZ4 A0A2A3EK09 A0A182GDP6 A0A1J1IEQ0 Q17DG0 A0A1I8MXF1 A0A0L0CI26 A0A1I8NQ95 T1PHM8 A0A034VCR1 A0A0M5L067 B4JV20 B3M1A8 B4LZW3 A0A182Y6H3 A0A1I8NQA2 A0A182SE67 A0A0V0GA99 B4NKT6 A0A182RBY9 A0A182M1X1 B4KDK1 A0A023F5E3 A0A0A1WWC2 A0A182XAS3 W8BIE7 A0A182UQB3 A0A182LEP3 A0A182U3D5 Q7QCE1 A0A182HSL9 A0A0Q9WF19 A0A182JSF2 A0A0Q9WV23 A0A2M4AQU9 A0A182F701 A0A1Q3FEZ8 A0A1B0C8D5 A0A0T6AW96 A0A182NZU9 A0A1Y1NDJ9 B4HE19 T1IBT2 A0A0M5J9F3 A0A1W4W0A6 A0A224X8N4 A0A182W0Y2 A0A0Q9WY51 A0A0Q9X5D7 A0A3B0JMS6 D2A654 A0A182QDI6 W5JVQ3 A0A182NRI9 A0A3B0JT03 A0A2P8XAU2 A0A3B0JRC6 B4QZ52 D5SHR2 B4PS90 B3P0Q7 Q9VFC8 A0A1W4VN51 A0A1Q3FEX8 B0WDW2 Q296I4 A0A3B0JMR5 U5ET57 A0A2H8TKY8 B4GF43 A0A0R1E850 A0A0R3NIA9 Q9VFC8-2
A0A154P9X4 A0A195C3G8 A0A195AVG9 A0A026X010 A0A195ETM8 A0A3L8E0G6 A0A2J7RI90 A0A151WGI9 A0A195E342 A0A2J7RI94 F4WA55 A0A0N0BF19 A0A067QZJ4 E2BGV0 A0A1B6CWL5 C0KJJ5 A0A088ARU9 E2A8G5 A0A1B6KQ54 A0A1B6GZ27 A0A0L7RDK2 E0VIZ4 A0A2A3EK09 A0A182GDP6 A0A1J1IEQ0 Q17DG0 A0A1I8MXF1 A0A0L0CI26 A0A1I8NQ95 T1PHM8 A0A034VCR1 A0A0M5L067 B4JV20 B3M1A8 B4LZW3 A0A182Y6H3 A0A1I8NQA2 A0A182SE67 A0A0V0GA99 B4NKT6 A0A182RBY9 A0A182M1X1 B4KDK1 A0A023F5E3 A0A0A1WWC2 A0A182XAS3 W8BIE7 A0A182UQB3 A0A182LEP3 A0A182U3D5 Q7QCE1 A0A182HSL9 A0A0Q9WF19 A0A182JSF2 A0A0Q9WV23 A0A2M4AQU9 A0A182F701 A0A1Q3FEZ8 A0A1B0C8D5 A0A0T6AW96 A0A182NZU9 A0A1Y1NDJ9 B4HE19 T1IBT2 A0A0M5J9F3 A0A1W4W0A6 A0A224X8N4 A0A182W0Y2 A0A0Q9WY51 A0A0Q9X5D7 A0A3B0JMS6 D2A654 A0A182QDI6 W5JVQ3 A0A182NRI9 A0A3B0JT03 A0A2P8XAU2 A0A3B0JRC6 B4QZ52 D5SHR2 B4PS90 B3P0Q7 Q9VFC8 A0A1W4VN51 A0A1Q3FEX8 B0WDW2 Q296I4 A0A3B0JMR5 U5ET57 A0A2H8TKY8 B4GF43 A0A0R1E850 A0A0R3NIA9 Q9VFC8-2
EC Number
2.4.1.11
Pubmed
19121390
26354079
22118469
24508170
30249741
21719571
+ More
24845553 20798317 20566863 26483478 17510324 25315136 26108605 25348373 17994087 25244985 25474469 25830018 24495485 20966253 12364791 14747013 17210077 28004739 18362917 19820115 20920257 23761445 29403074 17550304 10731132 12537572 12537569 17372656 18327897 24265594 15632085
24845553 20798317 20566863 26483478 17510324 25315136 26108605 25348373 17994087 25244985 25474469 25830018 24495485 20966253 12364791 14747013 17210077 28004739 18362917 19820115 20920257 23761445 29403074 17550304 10731132 12537572 12537569 17372656 18327897 24265594 15632085
EMBL
BABH01009723
BABH01009724
KQ459875
KPJ19685.1
NWSH01006076
PCG63683.1
+ More
ODYU01004881 SOQ45209.1 AGBW02012131 OWR45581.1 FJ809918 ADA53796.1 KQ434856 KZC08736.1 KQ978350 KYM94723.1 KQ976737 KYM75969.1 KK107069 EZA60734.1 KQ981975 KYN31605.1 QOIP01000002 RLU26147.1 NEVH01003505 PNF40545.1 KQ983167 KYQ46941.1 KQ979763 KYN19294.1 PNF40546.1 GL888041 EGI68911.1 KQ435814 KOX72681.1 KK852809 KDR15979.1 GL448228 EFN85082.1 GEDC01019480 GEDC01012728 JAS17818.1 JAS24570.1 FJ609646 ACM78946.1 GL437593 EFN70285.1 GEBQ01026407 JAT13570.1 GECZ01002111 JAS67658.1 KQ414614 KOC68865.1 DS235215 EEB13350.1 KZ288222 PBC32105.1 JXUM01010235 KQ560302 KXJ83162.1 CVRI01000048 CRK98691.1 CH477295 EAT44424.1 JRES01000344 KNC32063.1 KA647393 AFP62022.1 GAKP01018683 JAC40269.1 KR075826 ALE20545.1 CH916374 EDV91340.1 CH902617 EDV42135.1 CH940650 EDW67191.1 GECL01001052 JAP05072.1 CH964272 EDW84147.2 AXCM01000202 CH933806 EDW13835.1 GBBI01002358 JAC16354.1 GBXI01010958 JAD03334.1 GAMC01008058 JAB98497.1 AAAB01008859 EAA08045.3 APCN01000295 KRF83149.1 KRF99982.1 GGFK01009657 MBW42978.1 GFDL01008930 JAV26115.1 AJWK01000444 LJIG01022657 KRT79402.1 GEZM01005726 GEZM01005725 JAV95963.1 CH480815 EDW42111.1 ACPB03004993 CP012526 ALC46911.1 GFTR01007741 JAW08685.1 KRG00661.1 KRG00660.1 OUUW01000007 SPP83535.1 KQ971346 EFA05468.1 AXCN02001457 ADMH02000177 ETN67493.1 SPP83532.1 PYGN01006873 PSN29113.1 SPP83533.1 CM000364 EDX12880.1 BT124863 ADG03448.1 CM000160 EDW97510.1 CH954181 EDV48955.1 AE014297 AY052005 GFDL01008941 JAV26104.1 DS231902 EDS45043.1 CM000070 EAL28473.3 SPP83534.1 GANO01002077 JAB57794.1 GFXV01002875 MBW14680.1 CH479182 EDW34228.1 KRK03794.1 KRT00745.1
ODYU01004881 SOQ45209.1 AGBW02012131 OWR45581.1 FJ809918 ADA53796.1 KQ434856 KZC08736.1 KQ978350 KYM94723.1 KQ976737 KYM75969.1 KK107069 EZA60734.1 KQ981975 KYN31605.1 QOIP01000002 RLU26147.1 NEVH01003505 PNF40545.1 KQ983167 KYQ46941.1 KQ979763 KYN19294.1 PNF40546.1 GL888041 EGI68911.1 KQ435814 KOX72681.1 KK852809 KDR15979.1 GL448228 EFN85082.1 GEDC01019480 GEDC01012728 JAS17818.1 JAS24570.1 FJ609646 ACM78946.1 GL437593 EFN70285.1 GEBQ01026407 JAT13570.1 GECZ01002111 JAS67658.1 KQ414614 KOC68865.1 DS235215 EEB13350.1 KZ288222 PBC32105.1 JXUM01010235 KQ560302 KXJ83162.1 CVRI01000048 CRK98691.1 CH477295 EAT44424.1 JRES01000344 KNC32063.1 KA647393 AFP62022.1 GAKP01018683 JAC40269.1 KR075826 ALE20545.1 CH916374 EDV91340.1 CH902617 EDV42135.1 CH940650 EDW67191.1 GECL01001052 JAP05072.1 CH964272 EDW84147.2 AXCM01000202 CH933806 EDW13835.1 GBBI01002358 JAC16354.1 GBXI01010958 JAD03334.1 GAMC01008058 JAB98497.1 AAAB01008859 EAA08045.3 APCN01000295 KRF83149.1 KRF99982.1 GGFK01009657 MBW42978.1 GFDL01008930 JAV26115.1 AJWK01000444 LJIG01022657 KRT79402.1 GEZM01005726 GEZM01005725 JAV95963.1 CH480815 EDW42111.1 ACPB03004993 CP012526 ALC46911.1 GFTR01007741 JAW08685.1 KRG00661.1 KRG00660.1 OUUW01000007 SPP83535.1 KQ971346 EFA05468.1 AXCN02001457 ADMH02000177 ETN67493.1 SPP83532.1 PYGN01006873 PSN29113.1 SPP83533.1 CM000364 EDX12880.1 BT124863 ADG03448.1 CM000160 EDW97510.1 CH954181 EDV48955.1 AE014297 AY052005 GFDL01008941 JAV26104.1 DS231902 EDS45043.1 CM000070 EAL28473.3 SPP83534.1 GANO01002077 JAB57794.1 GFXV01002875 MBW14680.1 CH479182 EDW34228.1 KRK03794.1 KRT00745.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000076502
UP000078542
+ More
UP000078540 UP000053097 UP000078541 UP000279307 UP000235965 UP000075809 UP000078492 UP000007755 UP000053105 UP000027135 UP000008237 UP000005203 UP000000311 UP000053825 UP000009046 UP000242457 UP000069940 UP000249989 UP000183832 UP000008820 UP000095301 UP000037069 UP000095300 UP000001070 UP000007801 UP000008792 UP000076408 UP000075901 UP000007798 UP000075900 UP000075883 UP000009192 UP000076407 UP000075903 UP000075882 UP000075902 UP000007062 UP000075840 UP000075881 UP000069272 UP000092461 UP000075885 UP000001292 UP000015103 UP000092553 UP000192221 UP000075920 UP000268350 UP000007266 UP000075886 UP000000673 UP000075884 UP000245037 UP000000304 UP000002282 UP000008711 UP000000803 UP000002320 UP000001819 UP000008744
UP000078540 UP000053097 UP000078541 UP000279307 UP000235965 UP000075809 UP000078492 UP000007755 UP000053105 UP000027135 UP000008237 UP000005203 UP000000311 UP000053825 UP000009046 UP000242457 UP000069940 UP000249989 UP000183832 UP000008820 UP000095301 UP000037069 UP000095300 UP000001070 UP000007801 UP000008792 UP000076408 UP000075901 UP000007798 UP000075900 UP000075883 UP000009192 UP000076407 UP000075903 UP000075882 UP000075902 UP000007062 UP000075840 UP000075881 UP000069272 UP000092461 UP000075885 UP000001292 UP000015103 UP000092553 UP000192221 UP000075920 UP000268350 UP000007266 UP000075886 UP000000673 UP000075884 UP000245037 UP000000304 UP000002282 UP000008711 UP000000803 UP000002320 UP000001819 UP000008744
Pfam
Interpro
ProteinModelPortal
H9JPA6
A0A194RQV1
A0A2A4IX43
A0A2H1VWK1
A0A212EVT8
F2Q6J8
+ More
A0A154P9X4 A0A195C3G8 A0A195AVG9 A0A026X010 A0A195ETM8 A0A3L8E0G6 A0A2J7RI90 A0A151WGI9 A0A195E342 A0A2J7RI94 F4WA55 A0A0N0BF19 A0A067QZJ4 E2BGV0 A0A1B6CWL5 C0KJJ5 A0A088ARU9 E2A8G5 A0A1B6KQ54 A0A1B6GZ27 A0A0L7RDK2 E0VIZ4 A0A2A3EK09 A0A182GDP6 A0A1J1IEQ0 Q17DG0 A0A1I8MXF1 A0A0L0CI26 A0A1I8NQ95 T1PHM8 A0A034VCR1 A0A0M5L067 B4JV20 B3M1A8 B4LZW3 A0A182Y6H3 A0A1I8NQA2 A0A182SE67 A0A0V0GA99 B4NKT6 A0A182RBY9 A0A182M1X1 B4KDK1 A0A023F5E3 A0A0A1WWC2 A0A182XAS3 W8BIE7 A0A182UQB3 A0A182LEP3 A0A182U3D5 Q7QCE1 A0A182HSL9 A0A0Q9WF19 A0A182JSF2 A0A0Q9WV23 A0A2M4AQU9 A0A182F701 A0A1Q3FEZ8 A0A1B0C8D5 A0A0T6AW96 A0A182NZU9 A0A1Y1NDJ9 B4HE19 T1IBT2 A0A0M5J9F3 A0A1W4W0A6 A0A224X8N4 A0A182W0Y2 A0A0Q9WY51 A0A0Q9X5D7 A0A3B0JMS6 D2A654 A0A182QDI6 W5JVQ3 A0A182NRI9 A0A3B0JT03 A0A2P8XAU2 A0A3B0JRC6 B4QZ52 D5SHR2 B4PS90 B3P0Q7 Q9VFC8 A0A1W4VN51 A0A1Q3FEX8 B0WDW2 Q296I4 A0A3B0JMR5 U5ET57 A0A2H8TKY8 B4GF43 A0A0R1E850 A0A0R3NIA9 Q9VFC8-2
A0A154P9X4 A0A195C3G8 A0A195AVG9 A0A026X010 A0A195ETM8 A0A3L8E0G6 A0A2J7RI90 A0A151WGI9 A0A195E342 A0A2J7RI94 F4WA55 A0A0N0BF19 A0A067QZJ4 E2BGV0 A0A1B6CWL5 C0KJJ5 A0A088ARU9 E2A8G5 A0A1B6KQ54 A0A1B6GZ27 A0A0L7RDK2 E0VIZ4 A0A2A3EK09 A0A182GDP6 A0A1J1IEQ0 Q17DG0 A0A1I8MXF1 A0A0L0CI26 A0A1I8NQ95 T1PHM8 A0A034VCR1 A0A0M5L067 B4JV20 B3M1A8 B4LZW3 A0A182Y6H3 A0A1I8NQA2 A0A182SE67 A0A0V0GA99 B4NKT6 A0A182RBY9 A0A182M1X1 B4KDK1 A0A023F5E3 A0A0A1WWC2 A0A182XAS3 W8BIE7 A0A182UQB3 A0A182LEP3 A0A182U3D5 Q7QCE1 A0A182HSL9 A0A0Q9WF19 A0A182JSF2 A0A0Q9WV23 A0A2M4AQU9 A0A182F701 A0A1Q3FEZ8 A0A1B0C8D5 A0A0T6AW96 A0A182NZU9 A0A1Y1NDJ9 B4HE19 T1IBT2 A0A0M5J9F3 A0A1W4W0A6 A0A224X8N4 A0A182W0Y2 A0A0Q9WY51 A0A0Q9X5D7 A0A3B0JMS6 D2A654 A0A182QDI6 W5JVQ3 A0A182NRI9 A0A3B0JT03 A0A2P8XAU2 A0A3B0JRC6 B4QZ52 D5SHR2 B4PS90 B3P0Q7 Q9VFC8 A0A1W4VN51 A0A1Q3FEX8 B0WDW2 Q296I4 A0A3B0JMR5 U5ET57 A0A2H8TKY8 B4GF43 A0A0R1E850 A0A0R3NIA9 Q9VFC8-2
PDB
4QLB
E-value=1.26879e-111,
Score=1030
Ontologies
GO
PANTHER
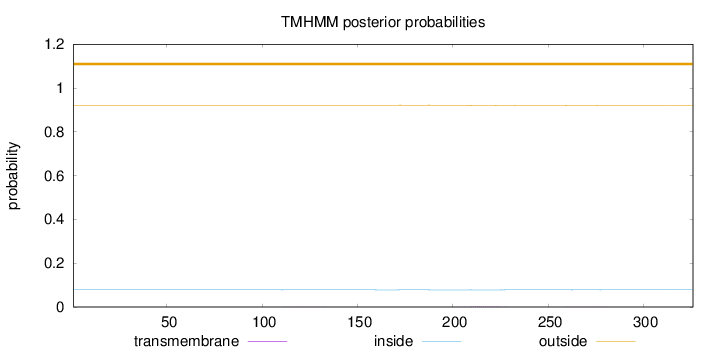
Topology
Subcellular location
Length:
326
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05076
Exp number, first 60 AAs:
0.0022
Total prob of N-in:
0.07869
outside
1 - 326
Population Genetic Test Statistics
Pi
226.380973
Theta
173.905088
Tajima's D
0.904812
CLR
0.015949
CSRT
0.631968401579921
Interpretation
Uncertain
