Gene
KWMTBOMO14027 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011246
Annotation
PREDICTED:_enhancer_of_rudimentary_homolog_[Plutella_xylostella]
Full name
Enhancer of rudimentary homolog
+ More
Protein enhancer of rudimentary
Protein enhancer of rudimentary
Location in the cell
Extracellular Reliability : 1.86 Nuclear Reliability : 1.588
Sequence
CDS
ATGAGTCACACAATTTTGCTGGTGCAGCCCGGTCCGAGGCCCGAAACCAGGACTTACTCAGACTACGAAAGCGTCAACGACTGCATGGAGGGTGTGTGTAAAATATATGAGGAACATCTGAAAAGGCGAAACCCTAATACACCCACTATCACGTACGATATATCACAACTCTTCGATTTTGTAGACCAGTTGGCAGATCTGAGCTGTTTAGTTTATCAGAAATCTACAAACACATACGCACCCTACAACAAAGATTGGATAAAGGAAAAGATATATGTTTTATTGCGTCAAGCGGCCGGACAGAGTGATTAA
Protein
MSHTILLVQPGPRPETRTYSDYESVNDCMEGVCKIYEEHLKRRNPNTPTITYDISQLFDFVDQLADLSCLVYQKSTNTYAPYNKDWIKEKIYVLLRQAAGQSD
Summary
Description
May have a role in the cell cycle.
Acts as an enhancer of the rudimentary gene. Has a role in pyrimidine biosynthesis and the cell cycle.
Acts as an enhancer of the rudimentary gene. Has a role in pyrimidine biosynthesis and the cell cycle.
Subunit
Homodimer.
Similarity
Belongs to the E(R) family.
Keywords
Cell cycle
Complete proteome
Reference proteome
Phosphoprotein
Pyrimidine biosynthesis
Feature
chain Enhancer of rudimentary homolog
Uniprot
A0A1E1WHB2
A0A2W1B8T9
A0A2A4J6T9
A0A2H1VWN5
I4DKE5
A0A194RU83
+ More
A0A194PU63 A0A0L7KMT8 A0A1L8DDD6 A0A026WDB9 A0A1B0D238 W8BT70 A0A034W7G7 A0A0C9R8S4 A0A0A1XGL2 A0A0K8V2C8 D2A5H4 A0A023EED8 Q93104 A0A0T6AWU7 A0A0K8TQT6 A0A1Y1NKQ2 A0A2M3ZCH8 A0A2M4AY89 A0A2M4C595 W5JTD2 A0A182FMK8 A0A087ZPW4 A0A195B7Q0 A0A158NVV4 E2BLS0 A0A3L8E2N7 A0A1W4XRE0 A0A310SIL9 A0A0L0C1B9 A0A0M8ZNJ2 A0A1I8Q0J0 A0A1Q3F692 A0A195FVL1 U5EVU7 B0X284 A0A1I8N100 A0A2M4C4I7 A0A182J3S3 F4WNR8 A0A0M3QZM7 A0A182TXD3 Q7QFJ0 A0A1I8JT96 A0A1A9YQM3 A0A1B0BIT6 A0A1A9VVA8 A0A1A9W6E6 A0A0P4WMY9 B3NUK0 B4Q002 A0A182M6T7 A0A182Y7T5 B4NW08 B4IJT8 M9PJE8 Q24337 A0A182Y7T6 A0A067QGF0 A0A2P8YY67 A0A1L8DAY8 B4L3J8 Q94554 D3TNV4 A0A182PNR2 A0A1W4UR18 B3MZI0 T1H439 C1C0S1 B4NPY5 A0A1B0FMW2 A0A3B0K7H1 B4GVI5 Q29GR5 A0A182XEG3 D3PJG3 C1BNA9 D3PGG7 A0A182W4K0 C1C2J2 B4JKD2 A0A3R7MIF4 E9HSC8 A0A0P5KF17 C1BNH7 A0A1A9Z9R0 A0A182L870 A0A182R9I1 C1BQF8 E0V9A8 C1C1R4 A0A336LFQ6 A0A0N8BQY3 A0A224Y1H8 A0A0V0GAG8
A0A194PU63 A0A0L7KMT8 A0A1L8DDD6 A0A026WDB9 A0A1B0D238 W8BT70 A0A034W7G7 A0A0C9R8S4 A0A0A1XGL2 A0A0K8V2C8 D2A5H4 A0A023EED8 Q93104 A0A0T6AWU7 A0A0K8TQT6 A0A1Y1NKQ2 A0A2M3ZCH8 A0A2M4AY89 A0A2M4C595 W5JTD2 A0A182FMK8 A0A087ZPW4 A0A195B7Q0 A0A158NVV4 E2BLS0 A0A3L8E2N7 A0A1W4XRE0 A0A310SIL9 A0A0L0C1B9 A0A0M8ZNJ2 A0A1I8Q0J0 A0A1Q3F692 A0A195FVL1 U5EVU7 B0X284 A0A1I8N100 A0A2M4C4I7 A0A182J3S3 F4WNR8 A0A0M3QZM7 A0A182TXD3 Q7QFJ0 A0A1I8JT96 A0A1A9YQM3 A0A1B0BIT6 A0A1A9VVA8 A0A1A9W6E6 A0A0P4WMY9 B3NUK0 B4Q002 A0A182M6T7 A0A182Y7T5 B4NW08 B4IJT8 M9PJE8 Q24337 A0A182Y7T6 A0A067QGF0 A0A2P8YY67 A0A1L8DAY8 B4L3J8 Q94554 D3TNV4 A0A182PNR2 A0A1W4UR18 B3MZI0 T1H439 C1C0S1 B4NPY5 A0A1B0FMW2 A0A3B0K7H1 B4GVI5 Q29GR5 A0A182XEG3 D3PJG3 C1BNA9 D3PGG7 A0A182W4K0 C1C2J2 B4JKD2 A0A3R7MIF4 E9HSC8 A0A0P5KF17 C1BNH7 A0A1A9Z9R0 A0A182L870 A0A182R9I1 C1BQF8 E0V9A8 C1C1R4 A0A336LFQ6 A0A0N8BQY3 A0A224Y1H8 A0A0V0GAG8
Pubmed
28756777
22651552
26354079
26227816
24508170
24495485
+ More
25348373 25830018 18362917 19820115 24945155 9074495 17510324 26369729 28004739 20920257 23761445 21347285 20798317 30249741 26108605 25315136 21719571 12364791 14747013 17210077 17994087 17550304 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7896098 12537569 24845553 29403074 20353571 15632085 23185243 21292972 20966253 20566863
25348373 25830018 18362917 19820115 24945155 9074495 17510324 26369729 28004739 20920257 23761445 21347285 20798317 30249741 26108605 25315136 21719571 12364791 14747013 17210077 17994087 17550304 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7896098 12537569 24845553 29403074 20353571 15632085 23185243 21292972 20966253 20566863
EMBL
GDQN01004661
JAT86393.1
KZ150262
PZC71728.1
NWSH01002975
PCG67160.1
+ More
ODYU01004881 SOQ45208.1 AK401763 BAM18385.1 KQ459875 KPJ19686.1 KQ459593 KPI96294.1 JTDY01008776 KOB64406.1 GFDF01009605 JAV04479.1 KK107260 EZA54062.1 AJVK01002660 GAMC01002010 JAC04546.1 GAKP01007416 JAC51536.1 GBYB01004555 JAG74322.1 GBXI01004211 JAD10081.1 GDHF01019308 GDHF01016895 GDHF01004980 JAI33006.1 JAI35419.1 JAI47334.1 KQ971345 EFA05372.1 GAPW01006387 GEHC01001475 JAC07211.1 JAV46170.1 U66869 CH477756 LJIG01022655 KRT79419.1 GDAI01001082 JAI16521.1 GEZM01001816 JAV97510.1 GGFM01005452 MBW26203.1 GGFK01012433 MBW45754.1 GGFJ01011017 MBW60158.1 ADMH02000428 ETN66543.1 KQ976565 KYM80553.1 ADTU01002904 GL449036 EFN83404.1 QOIP01000001 RLU26896.1 KQ760832 OAD58882.1 JRES01001135 KNC25249.1 KQ435966 KOX67920.1 GFDL01011978 JAV23067.1 KQ981215 KYN44498.1 GANO01000853 JAB59018.1 DS232282 EDS39107.1 GGFJ01011038 MBW60179.1 GL888237 EGI64261.1 CP012528 ALC49649.1 AAAB01008846 EAA06430.1 EAU77250.1 APCN01005666 JXJN01015174 GDRN01060176 JAI65413.1 CH954180 EDV46323.1 CM000162 EDX02208.1 AXCM01005429 CH991171 EDX16253.1 CH480849 EDW51241.1 AE014298 KX531563 AGB95206.1 AGB95207.1 ANY27373.1 L36921 BT001470 KK853541 KDR06873.1 PYGN01000290 PSN49187.1 GFDF01010557 JAV03527.1 CH933810 EDW07126.1 U66868 CH940660 EZ423106 ADD19382.1 CH902635 EDV33781.1 CAQQ02387213 BT080450 ACO14874.1 CH964291 EDW86210.1 CCAG010001225 OUUW01000015 SPP88622.1 CH479192 EDW26472.1 CH379064 EAL32044.2 BT121769 ADD38699.1 BT076088 BT076995 ACO10512.1 BT120723 ADD24363.1 BT081071 ACO15495.1 CH916370 EDW00035.1 QCYY01000803 ROT82567.1 GL732749 EFX65345.1 GDIQ01210814 LRGB01000243 JAK40911.1 KZS20219.1 BT076156 ACO10580.1 BT076837 ACO11261.1 DS234991 EEB09964.1 BT080793 ACO15217.1 UFQS01003394 UFQT01003394 SSX15597.1 SSX34959.1 GDIQ01153595 JAK98130.1 GFTR01001564 JAW14862.1 GECL01001312 JAP04812.1
ODYU01004881 SOQ45208.1 AK401763 BAM18385.1 KQ459875 KPJ19686.1 KQ459593 KPI96294.1 JTDY01008776 KOB64406.1 GFDF01009605 JAV04479.1 KK107260 EZA54062.1 AJVK01002660 GAMC01002010 JAC04546.1 GAKP01007416 JAC51536.1 GBYB01004555 JAG74322.1 GBXI01004211 JAD10081.1 GDHF01019308 GDHF01016895 GDHF01004980 JAI33006.1 JAI35419.1 JAI47334.1 KQ971345 EFA05372.1 GAPW01006387 GEHC01001475 JAC07211.1 JAV46170.1 U66869 CH477756 LJIG01022655 KRT79419.1 GDAI01001082 JAI16521.1 GEZM01001816 JAV97510.1 GGFM01005452 MBW26203.1 GGFK01012433 MBW45754.1 GGFJ01011017 MBW60158.1 ADMH02000428 ETN66543.1 KQ976565 KYM80553.1 ADTU01002904 GL449036 EFN83404.1 QOIP01000001 RLU26896.1 KQ760832 OAD58882.1 JRES01001135 KNC25249.1 KQ435966 KOX67920.1 GFDL01011978 JAV23067.1 KQ981215 KYN44498.1 GANO01000853 JAB59018.1 DS232282 EDS39107.1 GGFJ01011038 MBW60179.1 GL888237 EGI64261.1 CP012528 ALC49649.1 AAAB01008846 EAA06430.1 EAU77250.1 APCN01005666 JXJN01015174 GDRN01060176 JAI65413.1 CH954180 EDV46323.1 CM000162 EDX02208.1 AXCM01005429 CH991171 EDX16253.1 CH480849 EDW51241.1 AE014298 KX531563 AGB95206.1 AGB95207.1 ANY27373.1 L36921 BT001470 KK853541 KDR06873.1 PYGN01000290 PSN49187.1 GFDF01010557 JAV03527.1 CH933810 EDW07126.1 U66868 CH940660 EZ423106 ADD19382.1 CH902635 EDV33781.1 CAQQ02387213 BT080450 ACO14874.1 CH964291 EDW86210.1 CCAG010001225 OUUW01000015 SPP88622.1 CH479192 EDW26472.1 CH379064 EAL32044.2 BT121769 ADD38699.1 BT076088 BT076995 ACO10512.1 BT120723 ADD24363.1 BT081071 ACO15495.1 CH916370 EDW00035.1 QCYY01000803 ROT82567.1 GL732749 EFX65345.1 GDIQ01210814 LRGB01000243 JAK40911.1 KZS20219.1 BT076156 ACO10580.1 BT076837 ACO11261.1 DS234991 EEB09964.1 BT080793 ACO15217.1 UFQS01003394 UFQT01003394 SSX15597.1 SSX34959.1 GDIQ01153595 JAK98130.1 GFTR01001564 JAW14862.1 GECL01001312 JAP04812.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000037510
UP000053097
UP000092462
+ More
UP000007266 UP000008820 UP000000673 UP000069272 UP000005203 UP000078540 UP000005205 UP000008237 UP000279307 UP000192223 UP000037069 UP000053105 UP000095300 UP000078541 UP000002320 UP000095301 UP000075880 UP000007755 UP000092553 UP000075902 UP000007062 UP000075840 UP000092443 UP000092460 UP000078200 UP000091820 UP000008711 UP000002282 UP000075883 UP000076408 UP000000304 UP000001292 UP000000803 UP000027135 UP000245037 UP000009192 UP000008792 UP000075885 UP000192221 UP000007801 UP000015102 UP000007798 UP000092444 UP000268350 UP000008744 UP000001819 UP000076407 UP000075920 UP000001070 UP000283509 UP000000305 UP000076858 UP000092445 UP000075882 UP000075900 UP000009046
UP000007266 UP000008820 UP000000673 UP000069272 UP000005203 UP000078540 UP000005205 UP000008237 UP000279307 UP000192223 UP000037069 UP000053105 UP000095300 UP000078541 UP000002320 UP000095301 UP000075880 UP000007755 UP000092553 UP000075902 UP000007062 UP000075840 UP000092443 UP000092460 UP000078200 UP000091820 UP000008711 UP000002282 UP000075883 UP000076408 UP000000304 UP000001292 UP000000803 UP000027135 UP000245037 UP000009192 UP000008792 UP000075885 UP000192221 UP000007801 UP000015102 UP000007798 UP000092444 UP000268350 UP000008744 UP000001819 UP000076407 UP000075920 UP000001070 UP000283509 UP000000305 UP000076858 UP000092445 UP000075882 UP000075900 UP000009046
Pfam
PF01133 ER
SUPFAM
SSF143875
SSF143875
Gene 3D
ProteinModelPortal
A0A1E1WHB2
A0A2W1B8T9
A0A2A4J6T9
A0A2H1VWN5
I4DKE5
A0A194RU83
+ More
A0A194PU63 A0A0L7KMT8 A0A1L8DDD6 A0A026WDB9 A0A1B0D238 W8BT70 A0A034W7G7 A0A0C9R8S4 A0A0A1XGL2 A0A0K8V2C8 D2A5H4 A0A023EED8 Q93104 A0A0T6AWU7 A0A0K8TQT6 A0A1Y1NKQ2 A0A2M3ZCH8 A0A2M4AY89 A0A2M4C595 W5JTD2 A0A182FMK8 A0A087ZPW4 A0A195B7Q0 A0A158NVV4 E2BLS0 A0A3L8E2N7 A0A1W4XRE0 A0A310SIL9 A0A0L0C1B9 A0A0M8ZNJ2 A0A1I8Q0J0 A0A1Q3F692 A0A195FVL1 U5EVU7 B0X284 A0A1I8N100 A0A2M4C4I7 A0A182J3S3 F4WNR8 A0A0M3QZM7 A0A182TXD3 Q7QFJ0 A0A1I8JT96 A0A1A9YQM3 A0A1B0BIT6 A0A1A9VVA8 A0A1A9W6E6 A0A0P4WMY9 B3NUK0 B4Q002 A0A182M6T7 A0A182Y7T5 B4NW08 B4IJT8 M9PJE8 Q24337 A0A182Y7T6 A0A067QGF0 A0A2P8YY67 A0A1L8DAY8 B4L3J8 Q94554 D3TNV4 A0A182PNR2 A0A1W4UR18 B3MZI0 T1H439 C1C0S1 B4NPY5 A0A1B0FMW2 A0A3B0K7H1 B4GVI5 Q29GR5 A0A182XEG3 D3PJG3 C1BNA9 D3PGG7 A0A182W4K0 C1C2J2 B4JKD2 A0A3R7MIF4 E9HSC8 A0A0P5KF17 C1BNH7 A0A1A9Z9R0 A0A182L870 A0A182R9I1 C1BQF8 E0V9A8 C1C1R4 A0A336LFQ6 A0A0N8BQY3 A0A224Y1H8 A0A0V0GAG8
A0A194PU63 A0A0L7KMT8 A0A1L8DDD6 A0A026WDB9 A0A1B0D238 W8BT70 A0A034W7G7 A0A0C9R8S4 A0A0A1XGL2 A0A0K8V2C8 D2A5H4 A0A023EED8 Q93104 A0A0T6AWU7 A0A0K8TQT6 A0A1Y1NKQ2 A0A2M3ZCH8 A0A2M4AY89 A0A2M4C595 W5JTD2 A0A182FMK8 A0A087ZPW4 A0A195B7Q0 A0A158NVV4 E2BLS0 A0A3L8E2N7 A0A1W4XRE0 A0A310SIL9 A0A0L0C1B9 A0A0M8ZNJ2 A0A1I8Q0J0 A0A1Q3F692 A0A195FVL1 U5EVU7 B0X284 A0A1I8N100 A0A2M4C4I7 A0A182J3S3 F4WNR8 A0A0M3QZM7 A0A182TXD3 Q7QFJ0 A0A1I8JT96 A0A1A9YQM3 A0A1B0BIT6 A0A1A9VVA8 A0A1A9W6E6 A0A0P4WMY9 B3NUK0 B4Q002 A0A182M6T7 A0A182Y7T5 B4NW08 B4IJT8 M9PJE8 Q24337 A0A182Y7T6 A0A067QGF0 A0A2P8YY67 A0A1L8DAY8 B4L3J8 Q94554 D3TNV4 A0A182PNR2 A0A1W4UR18 B3MZI0 T1H439 C1C0S1 B4NPY5 A0A1B0FMW2 A0A3B0K7H1 B4GVI5 Q29GR5 A0A182XEG3 D3PJG3 C1BNA9 D3PGG7 A0A182W4K0 C1C2J2 B4JKD2 A0A3R7MIF4 E9HSC8 A0A0P5KF17 C1BNH7 A0A1A9Z9R0 A0A182L870 A0A182R9I1 C1BQF8 E0V9A8 C1C1R4 A0A336LFQ6 A0A0N8BQY3 A0A224Y1H8 A0A0V0GAG8
PDB
1WZ7
E-value=1.84575e-43,
Score=436
Ontologies
GO
PANTHER
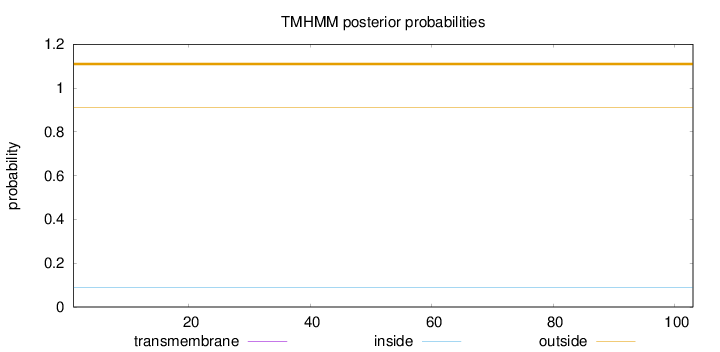
Topology
Length:
103
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00036
Exp number, first 60 AAs:
0.00013
Total prob of N-in:
0.08762
outside
1 - 103
Population Genetic Test Statistics
Pi
198.394613
Theta
185.350099
Tajima's D
0.016918
CLR
0.283596
CSRT
0.375581220938953
Interpretation
Uncertain
