Gene
KWMTBOMO14024 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011357
Annotation
PREDICTED:_DJ-1_beta_isoform_X1_[Bombyx_mori]
Full name
Protein dj-1beta
Location in the cell
Cytoplasmic Reliability : 0.824
Sequence
CDS
ATGAGCAAGTCTGCGTTAGTGATTTTGGCGCAGGGTGCAGAGGAGATGGAGACAGTTATAACTGTTGATATGCTGAGACGAGGAGGGGTAACAGTTACTCTAGCTGGCTTGGAAGGTTCATCTCCTGTGCTGTGCTCAAGGCAAGTGACTCTAGTACCTGATAAGTCACTGACTGAAGCCCTCGCTGAGAAACAACAGTATGATGCTGTGATTCTACCAGGAGGCTTAGAAGGATCAGACAGTCTCTCCAAATCCGAGAAGGTTGGTGCTCTGCTTAAAGACCACGAGGATAATGGGAAAATCATTGCTGCCATTTGTGCTGCTCCCATAGCGTTTGCAGCCCACGGCGTGGCGAGGGGCCGGCGCGTGACGTCATACCCCAGCACCAGGGACAAGCTGTCGGCCGGCGACTACACGTACGTGGAGGGGGAGAGGGTGGTGGTGGACGGCAACGTGGTCACCAGCCGGGGTCCCGGCACGGCGTACTGGTTTGGTTTAACCCTCATCGAGTTGCTGACGGGAAAGGAAAAGGCCGATCAAGTCGAAAAGGGAATGTTAATCTCGCAGTACTAA
Protein
MSKSALVILAQGAEEMETVITVDMLRRGGVTVTLAGLEGSSPVLCSRQVTLVPDKSLTEALAEKQQYDAVILPGGLEGSDSLSKSEKVGALLKDHEDNGKIIAAICAAPIAFAAHGVARGRRVTSYPSTRDKLSAGDYTYVEGERVVVDGNVVTSRGPGTAYWFGLTLIELLTGKEKADQVEKGMLISQY
Summary
Description
Involved in protection against reactive oxygen species (ROS) (By similarity). Does not play a role in methylglyoxal detoxification (PubMed:27903648).
Keywords
3D-structure
Alternative splicing
Complete proteome
Oxidation
Reference proteome
Feature
chain Protein dj-1beta
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JPA3
A0A194RIP6
S4PYG8
I4DP02
A0A194PS95
A0A3S2P3Q5
+ More
A0A212EVS5 A0A2A4IYT8 A0A2W1BCW7 A0A2W1BFE0 A0A293M4V2 A0A0K8RKN1 A0A1Z5L553 A0A0M4E692 A0A2R5LHG4 U5EU18 A0A1W7RAN9 A0A224Z6P2 E9H857 A0A023FV27 A0A131Z259 A0A023G846 A0A3S0ZM91 T1IZC7 A0A1E1X1C5 B4LJF4 A0A069DQN1 V4CQ47 A0A131XBT6 A0A0P5CKE1 A0A0P5VJC1 E0VNK6 A0A0K8V1W1 L7LY48 A0A2L2XV23 A0A0P5JVJ2 A0A0P6HBF2 A0A0P5L8G7 A0A034WLN6 A0A224XKQ6 A0A1W0X7Y1 A0A087UUC7 A0A0P5GFN8 A0A0B7BVV6 B4JW21 A0A3B0KBB7 A0A2T7PPS7 B4KTB4 A0A0L0BP31 C4N1A5 A0A0V0G3L0 A0A1I8P0Z0 T1PFP0 A0A0A1WPE4 W8CEF4 A0A170Z0F2 B3MT79 A0A2C9JDD2 Q9VA37 A0A1I8NIM1 A0A0P4VY04 Q29CB8 A0A158N7S9 A0A182DZN9 Q9VA37-2 B4GNB4 T1HZ41 B4HZV8 A6YPG8 A0A0K8TQW7 A0A1L8DG54 B4N5V8 A0A1L8DGH5 A0A1B6FW73 A0A1Y1L0F5 A0A1W4U6B9 A0A1B6HJ61 A0A183HAN3 A0A0L7REP2 A0A1B6F9S9 B4PML8 B3P7W3 B4R2A1 J9EJ64 R4FM64 A0A3P6TIW5 A0A0R3RSL2 A0A0K8T5X3 A7SJ12 A0A0K8SSL6 A0A1I8ESL3 A0A0H5S602 A0A1B0DQA6 A0A1W4XB01 A0A1Q3FZ82 A0A0A7HPN7 A0A1L8EBB6 K4GJK3 T1L2V0 A0A1B6E1Q3
A0A212EVS5 A0A2A4IYT8 A0A2W1BCW7 A0A2W1BFE0 A0A293M4V2 A0A0K8RKN1 A0A1Z5L553 A0A0M4E692 A0A2R5LHG4 U5EU18 A0A1W7RAN9 A0A224Z6P2 E9H857 A0A023FV27 A0A131Z259 A0A023G846 A0A3S0ZM91 T1IZC7 A0A1E1X1C5 B4LJF4 A0A069DQN1 V4CQ47 A0A131XBT6 A0A0P5CKE1 A0A0P5VJC1 E0VNK6 A0A0K8V1W1 L7LY48 A0A2L2XV23 A0A0P5JVJ2 A0A0P6HBF2 A0A0P5L8G7 A0A034WLN6 A0A224XKQ6 A0A1W0X7Y1 A0A087UUC7 A0A0P5GFN8 A0A0B7BVV6 B4JW21 A0A3B0KBB7 A0A2T7PPS7 B4KTB4 A0A0L0BP31 C4N1A5 A0A0V0G3L0 A0A1I8P0Z0 T1PFP0 A0A0A1WPE4 W8CEF4 A0A170Z0F2 B3MT79 A0A2C9JDD2 Q9VA37 A0A1I8NIM1 A0A0P4VY04 Q29CB8 A0A158N7S9 A0A182DZN9 Q9VA37-2 B4GNB4 T1HZ41 B4HZV8 A6YPG8 A0A0K8TQW7 A0A1L8DG54 B4N5V8 A0A1L8DGH5 A0A1B6FW73 A0A1Y1L0F5 A0A1W4U6B9 A0A1B6HJ61 A0A183HAN3 A0A0L7REP2 A0A1B6F9S9 B4PML8 B3P7W3 B4R2A1 J9EJ64 R4FM64 A0A3P6TIW5 A0A0R3RSL2 A0A0K8T5X3 A7SJ12 A0A0K8SSL6 A0A1I8ESL3 A0A0H5S602 A0A1B0DQA6 A0A1W4XB01 A0A1Q3FZ82 A0A0A7HPN7 A0A1L8EBB6 K4GJK3 T1L2V0 A0A1B6E1Q3
Pubmed
19121390
21455296
26354079
23622113
22651552
22118469
+ More
28756777 28528879 28797301 21292972 26830274 28503490 17994087 26334808 23254933 28049606 20566863 25576852 26561354 25348373 26108605 19576987 25830018 24495485 15562597 15976810 10731132 12537572 12537569 16139213 27903648 22515803 25315136 27129103 15632085 22919073 18207082 26369729 28004739 17550304 17615350 26850696 17885136 23056606
28756777 28528879 28797301 21292972 26830274 28503490 17994087 26334808 23254933 28049606 20566863 25576852 26561354 25348373 26108605 19576987 25830018 24495485 15562597 15976810 10731132 12537572 12537569 16139213 27903648 22515803 25315136 27129103 15632085 22919073 18207082 26369729 28004739 17550304 17615350 26850696 17885136 23056606
EMBL
BABH01009719
AB281053
BAJ04328.1
KQ460152
KPJ17432.1
GAIX01003313
+ More
JAA89247.1 AK403223 BAM19642.1 KQ459593 KPI96311.1 RSAL01000031 RVE51598.1 AGBW02012131 OWR45586.1 NWSH01005363 NWSH01004294 PCG64273.1 PCG65264.1 KZ150262 PZC71725.1 KZ150689 PZC70383.1 GFWV01010392 MAA35121.1 GADI01002163 JAA71645.1 GFJQ02004642 JAW02328.1 CP012524 ALC41954.1 GGLE01004762 MBY08888.1 GANO01001674 JAB58197.1 GFAH01000181 JAV48208.1 GFPF01010938 MAA22084.1 GL732603 EFX72014.1 GBBL01002800 JAC24520.1 GEDV01003727 JAP84830.1 GBBM01006410 JAC29008.1 RQTK01000360 RUS81019.1 AFFK01020433 GFAC01006342 JAT92846.1 CH940648 EDW60534.2 GBGD01002962 JAC85927.1 KB199699 ESP04580.1 GEFH01004983 JAP63598.1 GDIP01170200 LRGB01000005 JAJ53202.1 KZS22033.1 GDIQ01186732 GDIP01099165 JAK64993.1 JAM04550.1 DS235340 EEB14962.1 GDHF01019405 JAI32909.1 GACK01008028 JAA57006.1 IAAA01008366 IAAA01008367 IAAA01008368 LAA00006.1 GDIQ01219024 JAK32701.1 GDIQ01021566 JAN73171.1 GDIQ01173334 JAK78391.1 GAKP01002461 JAC56491.1 GFTR01003361 JAW13065.1 MTYJ01000011 OQV23422.1 KK121650 KFM80966.1 GDIQ01247466 JAK04259.1 HACG01049606 CEK96471.1 CH916375 EDV98159.1 OUUW01000005 SPP80868.1 PZQS01000002 PVD35434.1 CH933808 EDW10626.1 JRES01001582 KNC21766.1 EZ048878 ACN69170.1 GECL01003491 JAP02633.1 KA647479 AFP62108.1 GBXI01013736 JAD00556.1 GAMC01000291 JAC06265.1 GEMB01002755 JAS00441.1 CH902623 EDV30469.2 AB079599 AE014297 AY060670 BAB84672.1 GDKW01002127 JAI54468.1 CM000070 EAL26728.2 CMVM020000376 UYRW01000136 VDK63568.1 CH479186 EDW39240.1 ACPB03004679 CH480819 EDW53565.1 EF638979 ABR27864.1 GDAI01000846 JAI16757.1 GFDF01008651 JAV05433.1 CH964154 EDW79747.2 GFDF01008650 JAV05434.1 GECZ01015313 JAS54456.1 GEZM01068147 GEZM01068146 JAV67142.1 GECU01032963 JAS74743.1 UZAJ01003496 VDO40290.1 KQ414608 KOC69422.1 GECZ01022822 JAS46947.1 CM000160 EDW99152.2 CH954182 EDV53021.1 CM000364 EDX15057.1 ADBV01002836 UYWW01001951 EJW82496.1 VDM11203.1 GAHY01001839 JAA75671.1 UYRX01001077 VDK88062.1 GBRD01004882 JAG60939.1 DS469673 EDO36318.1 GBRD01009660 JAG56164.1 LN856934 CRZ24035.1 AJVK01018992 GFDL01002177 JAV32868.1 KM408469 AIZ07943.1 GFDG01002894 JAV15905.1 JX052846 JX209533 AFK11074.1 AFM87847.1 CAEY01000991 GEDC01005442 JAS31856.1
JAA89247.1 AK403223 BAM19642.1 KQ459593 KPI96311.1 RSAL01000031 RVE51598.1 AGBW02012131 OWR45586.1 NWSH01005363 NWSH01004294 PCG64273.1 PCG65264.1 KZ150262 PZC71725.1 KZ150689 PZC70383.1 GFWV01010392 MAA35121.1 GADI01002163 JAA71645.1 GFJQ02004642 JAW02328.1 CP012524 ALC41954.1 GGLE01004762 MBY08888.1 GANO01001674 JAB58197.1 GFAH01000181 JAV48208.1 GFPF01010938 MAA22084.1 GL732603 EFX72014.1 GBBL01002800 JAC24520.1 GEDV01003727 JAP84830.1 GBBM01006410 JAC29008.1 RQTK01000360 RUS81019.1 AFFK01020433 GFAC01006342 JAT92846.1 CH940648 EDW60534.2 GBGD01002962 JAC85927.1 KB199699 ESP04580.1 GEFH01004983 JAP63598.1 GDIP01170200 LRGB01000005 JAJ53202.1 KZS22033.1 GDIQ01186732 GDIP01099165 JAK64993.1 JAM04550.1 DS235340 EEB14962.1 GDHF01019405 JAI32909.1 GACK01008028 JAA57006.1 IAAA01008366 IAAA01008367 IAAA01008368 LAA00006.1 GDIQ01219024 JAK32701.1 GDIQ01021566 JAN73171.1 GDIQ01173334 JAK78391.1 GAKP01002461 JAC56491.1 GFTR01003361 JAW13065.1 MTYJ01000011 OQV23422.1 KK121650 KFM80966.1 GDIQ01247466 JAK04259.1 HACG01049606 CEK96471.1 CH916375 EDV98159.1 OUUW01000005 SPP80868.1 PZQS01000002 PVD35434.1 CH933808 EDW10626.1 JRES01001582 KNC21766.1 EZ048878 ACN69170.1 GECL01003491 JAP02633.1 KA647479 AFP62108.1 GBXI01013736 JAD00556.1 GAMC01000291 JAC06265.1 GEMB01002755 JAS00441.1 CH902623 EDV30469.2 AB079599 AE014297 AY060670 BAB84672.1 GDKW01002127 JAI54468.1 CM000070 EAL26728.2 CMVM020000376 UYRW01000136 VDK63568.1 CH479186 EDW39240.1 ACPB03004679 CH480819 EDW53565.1 EF638979 ABR27864.1 GDAI01000846 JAI16757.1 GFDF01008651 JAV05433.1 CH964154 EDW79747.2 GFDF01008650 JAV05434.1 GECZ01015313 JAS54456.1 GEZM01068147 GEZM01068146 JAV67142.1 GECU01032963 JAS74743.1 UZAJ01003496 VDO40290.1 KQ414608 KOC69422.1 GECZ01022822 JAS46947.1 CM000160 EDW99152.2 CH954182 EDV53021.1 CM000364 EDX15057.1 ADBV01002836 UYWW01001951 EJW82496.1 VDM11203.1 GAHY01001839 JAA75671.1 UYRX01001077 VDK88062.1 GBRD01004882 JAG60939.1 DS469673 EDO36318.1 GBRD01009660 JAG56164.1 LN856934 CRZ24035.1 AJVK01018992 GFDL01002177 JAV32868.1 KM408469 AIZ07943.1 GFDG01002894 JAV15905.1 JX052846 JX209533 AFK11074.1 AFM87847.1 CAEY01000991 GEDC01005442 JAS31856.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000218220
+ More
UP000092553 UP000000305 UP000271974 UP000008792 UP000030746 UP000076858 UP000009046 UP000054359 UP000001070 UP000268350 UP000245119 UP000009192 UP000037069 UP000095300 UP000007801 UP000076420 UP000000803 UP000095301 UP000001819 UP000024404 UP000077448 UP000271087 UP000008744 UP000015103 UP000001292 UP000007798 UP000192221 UP000050787 UP000053825 UP000002282 UP000008711 UP000000304 UP000004810 UP000270924 UP000277928 UP000050640 UP000001593 UP000093561 UP000006672 UP000092462 UP000192223 UP000015104
UP000092553 UP000000305 UP000271974 UP000008792 UP000030746 UP000076858 UP000009046 UP000054359 UP000001070 UP000268350 UP000245119 UP000009192 UP000037069 UP000095300 UP000007801 UP000076420 UP000000803 UP000095301 UP000001819 UP000024404 UP000077448 UP000271087 UP000008744 UP000015103 UP000001292 UP000007798 UP000192221 UP000050787 UP000053825 UP000002282 UP000008711 UP000000304 UP000004810 UP000270924 UP000277928 UP000050640 UP000001593 UP000093561 UP000006672 UP000092462 UP000192223 UP000015104
Pfam
PF01965 DJ-1_PfpI
SUPFAM
SSF52317
SSF52317
Gene 3D
ProteinModelPortal
H9JPA3
A0A194RIP6
S4PYG8
I4DP02
A0A194PS95
A0A3S2P3Q5
+ More
A0A212EVS5 A0A2A4IYT8 A0A2W1BCW7 A0A2W1BFE0 A0A293M4V2 A0A0K8RKN1 A0A1Z5L553 A0A0M4E692 A0A2R5LHG4 U5EU18 A0A1W7RAN9 A0A224Z6P2 E9H857 A0A023FV27 A0A131Z259 A0A023G846 A0A3S0ZM91 T1IZC7 A0A1E1X1C5 B4LJF4 A0A069DQN1 V4CQ47 A0A131XBT6 A0A0P5CKE1 A0A0P5VJC1 E0VNK6 A0A0K8V1W1 L7LY48 A0A2L2XV23 A0A0P5JVJ2 A0A0P6HBF2 A0A0P5L8G7 A0A034WLN6 A0A224XKQ6 A0A1W0X7Y1 A0A087UUC7 A0A0P5GFN8 A0A0B7BVV6 B4JW21 A0A3B0KBB7 A0A2T7PPS7 B4KTB4 A0A0L0BP31 C4N1A5 A0A0V0G3L0 A0A1I8P0Z0 T1PFP0 A0A0A1WPE4 W8CEF4 A0A170Z0F2 B3MT79 A0A2C9JDD2 Q9VA37 A0A1I8NIM1 A0A0P4VY04 Q29CB8 A0A158N7S9 A0A182DZN9 Q9VA37-2 B4GNB4 T1HZ41 B4HZV8 A6YPG8 A0A0K8TQW7 A0A1L8DG54 B4N5V8 A0A1L8DGH5 A0A1B6FW73 A0A1Y1L0F5 A0A1W4U6B9 A0A1B6HJ61 A0A183HAN3 A0A0L7REP2 A0A1B6F9S9 B4PML8 B3P7W3 B4R2A1 J9EJ64 R4FM64 A0A3P6TIW5 A0A0R3RSL2 A0A0K8T5X3 A7SJ12 A0A0K8SSL6 A0A1I8ESL3 A0A0H5S602 A0A1B0DQA6 A0A1W4XB01 A0A1Q3FZ82 A0A0A7HPN7 A0A1L8EBB6 K4GJK3 T1L2V0 A0A1B6E1Q3
A0A212EVS5 A0A2A4IYT8 A0A2W1BCW7 A0A2W1BFE0 A0A293M4V2 A0A0K8RKN1 A0A1Z5L553 A0A0M4E692 A0A2R5LHG4 U5EU18 A0A1W7RAN9 A0A224Z6P2 E9H857 A0A023FV27 A0A131Z259 A0A023G846 A0A3S0ZM91 T1IZC7 A0A1E1X1C5 B4LJF4 A0A069DQN1 V4CQ47 A0A131XBT6 A0A0P5CKE1 A0A0P5VJC1 E0VNK6 A0A0K8V1W1 L7LY48 A0A2L2XV23 A0A0P5JVJ2 A0A0P6HBF2 A0A0P5L8G7 A0A034WLN6 A0A224XKQ6 A0A1W0X7Y1 A0A087UUC7 A0A0P5GFN8 A0A0B7BVV6 B4JW21 A0A3B0KBB7 A0A2T7PPS7 B4KTB4 A0A0L0BP31 C4N1A5 A0A0V0G3L0 A0A1I8P0Z0 T1PFP0 A0A0A1WPE4 W8CEF4 A0A170Z0F2 B3MT79 A0A2C9JDD2 Q9VA37 A0A1I8NIM1 A0A0P4VY04 Q29CB8 A0A158N7S9 A0A182DZN9 Q9VA37-2 B4GNB4 T1HZ41 B4HZV8 A6YPG8 A0A0K8TQW7 A0A1L8DG54 B4N5V8 A0A1L8DGH5 A0A1B6FW73 A0A1Y1L0F5 A0A1W4U6B9 A0A1B6HJ61 A0A183HAN3 A0A0L7REP2 A0A1B6F9S9 B4PML8 B3P7W3 B4R2A1 J9EJ64 R4FM64 A0A3P6TIW5 A0A0R3RSL2 A0A0K8T5X3 A7SJ12 A0A0K8SSL6 A0A1I8ESL3 A0A0H5S602 A0A1B0DQA6 A0A1W4XB01 A0A1Q3FZ82 A0A0A7HPN7 A0A1L8EBB6 K4GJK3 T1L2V0 A0A1B6E1Q3
PDB
4E08
E-value=1.17719e-41,
Score=423
Ontologies
GO
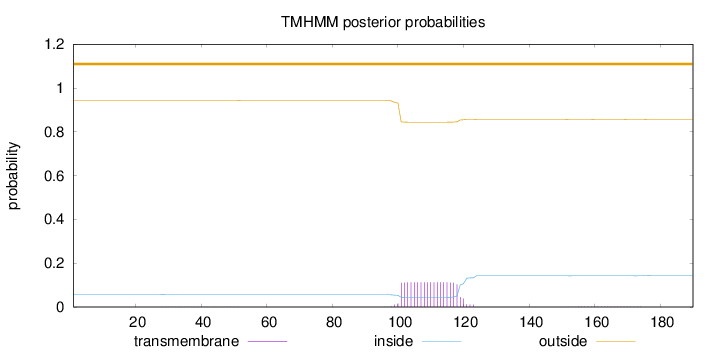
Topology
Length:
190
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.21006
Exp number, first 60 AAs:
0.01776
Total prob of N-in:
0.05712
outside
1 - 190
Population Genetic Test Statistics
Pi
178.794699
Theta
178.674205
Tajima's D
-0.217905
CLR
1.28138
CSRT
0.311284435778211
Interpretation
Uncertain
