Gene
KWMTBOMO14015
Annotation
PREDICTED:_ankyrin-3-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.916
Sequence
CDS
ATGCCTTCAGAGGTGATATCGGATAAGAGCTTGCAGAGGGAGCTGGCGGACTCCATTATCAGGATGGTGCCTTTGGATGAGATAAGGATCCTCCTGGCTTGTGGAGCCAAGGTGAATGAGCCCGTGACCCAAGGTCTCCGTCCGCTGCATTACGCGATCTGGCAGCGACACGTGGAAGCCACAAGGCTGCTCCTCGTCAGGGGCTGCGACGTCAACGCGACCGACGACTGCGGATACAGCGCCCTGCACCTCTCTGCTGAGCACGGGTACACGGAGCTGGTGAAGCTGCTACTGGAGAGCGGTGCAGCCGTGGACTACAGGCCCGACACCGGCGAAGAGTTCCCGAGGACGACGCTCTGTGACGAGCCGCTGCGACTGGCCATCAGGAACAAGCACTATGATGTTGCTAAGCTACTGCTGGAGCACGGCGCTGATCCAAACAAAAGGTACTTCTTCGGCTCCGAAATCAATCTGGTCTCCGACCCCGAGTACTTGGAGTTGCTGTTGACTTTCGGAGCCAACCCTGACTCCAGGGACCGAGCGGGGCTTACTCCTCTCATGAAAGCTGCTAGACAAAAGAAAGGTATAGAAGCAGTTCTGCTCCTCATCAGTTTCGGAGCTGATATCAACGCGATGGCCGACGCCAGGAACGACTACAGGACCGTCCTACACTACGCCGTGCTCAGTGGTAACCCAGATGTAGTGAACCTCATGATCAAGCAAGGAGCTCGCGTGAACTACGACTGTCCTGAACTCAGCAAGCCGAGCCCTCTGGACCTGGCCATCCTGAAGGGAGACGTGACCATGGTACAACTGCTCCTATCAGCTGGCGCTCAAGTGAACGCGTCTAGTTCCGTGATCGGGTCCCCGCTTCACGTCGCCTGCTCCGACAACATAGACAACAGAAAGGAAATCGTTAAAATCCTGCTGGAGAGCGGCGCCGACCCGAACCTGAAGGTGTACAGCCCGGAGGACGCGGCTCAGCTGCGGCCCGCGCTCGCCGAGTACCTCGCCAGCAACGTGCGGCCCCACCGCGACCTGGTGGCCATGCTCCTCGCGCACGGAGCCAGGGTCATAATGAAAACCCAGTTTCGTGATCCGGATGGCATTCTCAACCACTTACAAAACGTTACTTCAGAGGAATACGAGCATATCTTCTACTTACTTCTGGAAGCCGCAGAAGCCTTCGACCTCTGCATGATCAAGAGGAACAGTATATTAAACGCAGAACAAAAGCAAAAACTAATTGAACGCGCCAAAAATCCAATATCCCTATTGGCACAAGTCAGGATATTCCTCCGGAAACAGTTCGGCCCCTCCTTCCCCCACGCGGTCAAAACTATGGAAATACCTAAAACTCTTCACCACTACCTGCTTTTTGAATGTCGTTAA
Protein
MPSEVISDKSLQRELADSIIRMVPLDEIRILLACGAKVNEPVTQGLRPLHYAIWQRHVEATRLLLVRGCDVNATDDCGYSALHLSAEHGYTELVKLLLESGAAVDYRPDTGEEFPRTTLCDEPLRLAIRNKHYDVAKLLLEHGADPNKRYFFGSEINLVSDPEYLELLLTFGANPDSRDRAGLTPLMKAARQKKGIEAVLLLISFGADINAMADARNDYRTVLHYAVLSGNPDVVNLMIKQGARVNYDCPELSKPSPLDLAILKGDVTMVQLLLSAGAQVNASSSVIGSPLHVACSDNIDNRKEIVKILLESGADPNLKVYSPEDAAQLRPALAEYLASNVRPHRDLVAMLLAHGARVIMKTQFRDPDGILNHLQNVTSEEYEHIFYLLLEAAEAFDLCMIKRNSILNAEQKQKLIERAKNPISLLAQVRIFLRKQFGPSFPHAVKTMEIPKTLHHYLLFECR
Summary
Uniprot
A0A2A4JMK8
H9JNZ8
A0A2H1VSH3
A0A212EWP5
A0A194PTK0
A0A1Y1LM47
+ More
A0A0T6AZF4 A0A2J7QNA9 A0A1Y1LH82 U4UID2 A0A2P8XSC0 E0VA31 A0A195FXV0 A0A195C0Y5 A0A0M8ZME8 F4WJS3 A0A195BBC1 A0A154P9R3 A0A158NS50 A0A195DE68 E9IFP0 A0A151WH11 A0A3L8DE59 A0A0L7RD67 A0A087ZNK0 E2B8V5 A0A026VZC8 A0A0J7LAH5 A0A2W1BA10 A0A067QY70 A0A1B6E7D8 A0A0C9R3L5 A0A0C9QDM1 T1HSD4 A0A1J1I7R4 A0A023EY15 A0A1L8DJ79 A0A1B6LDD1 A0A1B6HYL7 A0A1Q3FNA6 A0A1Q3FNB6 B0W4J7 Q17K38 A0A1S4EZV1 A0A182G3H7 A0A2M4CFV6 W5JUE7 A0A182LZ69 A0A182IXN2 A0A1S4GY38 Q7Q4G9 A0A182PLS2 A0A182UHU4 A0A182IC57 A0A182VDF7 A0A2C9GNI8 A0A182Q9Z8 A0A182YDT6 A0A182W6F0 A0A182K1H6 A0A3F2Z0M9 A0A182NU05 A0A2R7W2B1 A0A084WH64 A0A1D2NFU2 A0A226D097 A0A182RDS2 A0A182F3G5 A0A0N8D0C4 A0A0P5DYT9 A0A0N8A3N1 A0A0P4ZSJ2 A0A0P5TQ10 A0A0P5P992 A0A0P5YX33 E9G0L5 A0A0N8D187 A0A0N7ZLN1 A0A0P5CC42 A0A0P5TEE5 T1IPD0 A0A0P6I067 A0A2L2YKQ9 A0A087V0C8 A0A194RHY2 A0A0P5KHA2 A0A139WG98 A0A0P5QD11 A0A0P6BES3 T1DCA4 A0A0P5I8A5 A0A0P5WWX7 A0A0P5A7C5 A0A0P5I988 A0A0P5I4W4 A0A0P5QR85 A0A0P6F0X6
A0A0T6AZF4 A0A2J7QNA9 A0A1Y1LH82 U4UID2 A0A2P8XSC0 E0VA31 A0A195FXV0 A0A195C0Y5 A0A0M8ZME8 F4WJS3 A0A195BBC1 A0A154P9R3 A0A158NS50 A0A195DE68 E9IFP0 A0A151WH11 A0A3L8DE59 A0A0L7RD67 A0A087ZNK0 E2B8V5 A0A026VZC8 A0A0J7LAH5 A0A2W1BA10 A0A067QY70 A0A1B6E7D8 A0A0C9R3L5 A0A0C9QDM1 T1HSD4 A0A1J1I7R4 A0A023EY15 A0A1L8DJ79 A0A1B6LDD1 A0A1B6HYL7 A0A1Q3FNA6 A0A1Q3FNB6 B0W4J7 Q17K38 A0A1S4EZV1 A0A182G3H7 A0A2M4CFV6 W5JUE7 A0A182LZ69 A0A182IXN2 A0A1S4GY38 Q7Q4G9 A0A182PLS2 A0A182UHU4 A0A182IC57 A0A182VDF7 A0A2C9GNI8 A0A182Q9Z8 A0A182YDT6 A0A182W6F0 A0A182K1H6 A0A3F2Z0M9 A0A182NU05 A0A2R7W2B1 A0A084WH64 A0A1D2NFU2 A0A226D097 A0A182RDS2 A0A182F3G5 A0A0N8D0C4 A0A0P5DYT9 A0A0N8A3N1 A0A0P4ZSJ2 A0A0P5TQ10 A0A0P5P992 A0A0P5YX33 E9G0L5 A0A0N8D187 A0A0N7ZLN1 A0A0P5CC42 A0A0P5TEE5 T1IPD0 A0A0P6I067 A0A2L2YKQ9 A0A087V0C8 A0A194RHY2 A0A0P5KHA2 A0A139WG98 A0A0P5QD11 A0A0P6BES3 T1DCA4 A0A0P5I8A5 A0A0P5WWX7 A0A0P5A7C5 A0A0P5I988 A0A0P5I4W4 A0A0P5QR85 A0A0P6F0X6
Pubmed
EMBL
NWSH01000978
PCG73287.1
BABH01009707
ODYU01004178
SOQ43770.1
AGBW02011924
+ More
OWR45903.1 KQ459593 KPI96303.1 GEZM01055645 JAV72995.1 LJIG01022455 KRT80502.1 NEVH01013198 PNF30058.1 GEZM01055646 JAV72994.1 KB632310 ERL92123.1 PYGN01001429 PSN34909.1 DS235004 EEB10237.1 KQ981208 KYN44669.1 KQ978457 KYM93833.1 KQ438551 KOX67195.1 GL888186 EGI65565.1 KQ976528 KYM81851.1 KQ434856 KZC08676.1 ADTU01024470 ADTU01024471 KQ980989 KYN10724.1 GL762880 EFZ20619.1 KQ983136 KYQ47095.1 QOIP01000010 RLU18179.1 KQ414614 KOC68803.1 GL446384 EFN87895.1 KK107661 EZA48224.1 LBMM01000111 KMR04892.1 KZ150299 PZC71541.1 KK852835 KDR15264.1 GEDC01010123 GEDC01003447 JAS27175.1 JAS33851.1 GBYB01001411 JAG71178.1 GBYB01001409 JAG71176.1 ACPB03004812 CVRI01000038 CRK94441.1 GBBI01004786 JAC13926.1 GFDF01007690 JAV06394.1 GEBQ01018286 GEBQ01010490 GEBQ01002167 JAT21691.1 JAT29487.1 JAT37810.1 GECU01027928 JAS79778.1 GFDL01005966 JAV29079.1 GFDL01006021 JAV29024.1 DS231838 EDS33842.1 CH477228 EAT47054.1 JXUM01141503 KQ569258 KXJ68695.1 GGFL01000025 MBW64203.1 ADMH02000463 ETN66379.1 AXCM01003506 AAAB01008964 EAA12523.4 APCN01000775 AXCN02000808 KK854262 PTY13887.1 ATLV01023791 KE525346 KFB49558.1 LJIJ01000062 ODN03826.1 LNIX01000048 OXA38081.1 GDIP01081255 JAM22460.1 GDIP01149805 LRGB01001581 JAJ73597.1 KZS11213.1 GDIP01185689 JAJ37713.1 GDIP01212334 JAJ11068.1 GDIP01126251 JAL77463.1 GDIQ01133848 JAL17878.1 GDIP01052102 JAM51613.1 GL732528 EFX86924.1 GDIP01078751 JAM24964.1 GDIP01233289 JAI90112.1 GDIP01188732 JAJ34670.1 GDIP01127645 JAL76069.1 JH431256 GDIQ01011366 JAN83371.1 IAAA01036158 LAA08577.1 KK122590 KFM83067.1 KQ460152 KPJ17438.1 GDIQ01184114 JAK67611.1 KQ971348 KYB26881.1 GDIQ01118900 JAL32826.1 GDIP01017268 JAM86447.1 GAKT01000097 JAA92965.1 GDIQ01218159 JAK33566.1 GDIP01081254 JAM22461.1 GDIP01203088 JAJ20314.1 GDIQ01217009 JAK34716.1 GDIQ01223500 JAK28225.1 GDIQ01110648 JAL41078.1 GDIQ01055864 JAN38873.1
OWR45903.1 KQ459593 KPI96303.1 GEZM01055645 JAV72995.1 LJIG01022455 KRT80502.1 NEVH01013198 PNF30058.1 GEZM01055646 JAV72994.1 KB632310 ERL92123.1 PYGN01001429 PSN34909.1 DS235004 EEB10237.1 KQ981208 KYN44669.1 KQ978457 KYM93833.1 KQ438551 KOX67195.1 GL888186 EGI65565.1 KQ976528 KYM81851.1 KQ434856 KZC08676.1 ADTU01024470 ADTU01024471 KQ980989 KYN10724.1 GL762880 EFZ20619.1 KQ983136 KYQ47095.1 QOIP01000010 RLU18179.1 KQ414614 KOC68803.1 GL446384 EFN87895.1 KK107661 EZA48224.1 LBMM01000111 KMR04892.1 KZ150299 PZC71541.1 KK852835 KDR15264.1 GEDC01010123 GEDC01003447 JAS27175.1 JAS33851.1 GBYB01001411 JAG71178.1 GBYB01001409 JAG71176.1 ACPB03004812 CVRI01000038 CRK94441.1 GBBI01004786 JAC13926.1 GFDF01007690 JAV06394.1 GEBQ01018286 GEBQ01010490 GEBQ01002167 JAT21691.1 JAT29487.1 JAT37810.1 GECU01027928 JAS79778.1 GFDL01005966 JAV29079.1 GFDL01006021 JAV29024.1 DS231838 EDS33842.1 CH477228 EAT47054.1 JXUM01141503 KQ569258 KXJ68695.1 GGFL01000025 MBW64203.1 ADMH02000463 ETN66379.1 AXCM01003506 AAAB01008964 EAA12523.4 APCN01000775 AXCN02000808 KK854262 PTY13887.1 ATLV01023791 KE525346 KFB49558.1 LJIJ01000062 ODN03826.1 LNIX01000048 OXA38081.1 GDIP01081255 JAM22460.1 GDIP01149805 LRGB01001581 JAJ73597.1 KZS11213.1 GDIP01185689 JAJ37713.1 GDIP01212334 JAJ11068.1 GDIP01126251 JAL77463.1 GDIQ01133848 JAL17878.1 GDIP01052102 JAM51613.1 GL732528 EFX86924.1 GDIP01078751 JAM24964.1 GDIP01233289 JAI90112.1 GDIP01188732 JAJ34670.1 GDIP01127645 JAL76069.1 JH431256 GDIQ01011366 JAN83371.1 IAAA01036158 LAA08577.1 KK122590 KFM83067.1 KQ460152 KPJ17438.1 GDIQ01184114 JAK67611.1 KQ971348 KYB26881.1 GDIQ01118900 JAL32826.1 GDIP01017268 JAM86447.1 GAKT01000097 JAA92965.1 GDIQ01218159 JAK33566.1 GDIP01081254 JAM22461.1 GDIP01203088 JAJ20314.1 GDIQ01217009 JAK34716.1 GDIQ01223500 JAK28225.1 GDIQ01110648 JAL41078.1 GDIQ01055864 JAN38873.1
Proteomes
UP000218220
UP000005204
UP000007151
UP000053268
UP000235965
UP000030742
+ More
UP000245037 UP000009046 UP000078541 UP000078542 UP000053105 UP000007755 UP000078540 UP000076502 UP000005205 UP000078492 UP000075809 UP000279307 UP000053825 UP000005203 UP000008237 UP000053097 UP000036403 UP000027135 UP000015103 UP000183832 UP000002320 UP000008820 UP000069940 UP000249989 UP000000673 UP000075883 UP000075880 UP000007062 UP000075885 UP000075902 UP000075840 UP000075903 UP000075886 UP000076408 UP000075920 UP000075881 UP000075884 UP000030765 UP000094527 UP000198287 UP000075900 UP000069272 UP000076858 UP000000305 UP000054359 UP000053240 UP000007266
UP000245037 UP000009046 UP000078541 UP000078542 UP000053105 UP000007755 UP000078540 UP000076502 UP000005205 UP000078492 UP000075809 UP000279307 UP000053825 UP000005203 UP000008237 UP000053097 UP000036403 UP000027135 UP000015103 UP000183832 UP000002320 UP000008820 UP000069940 UP000249989 UP000000673 UP000075883 UP000075880 UP000007062 UP000075885 UP000075902 UP000075840 UP000075903 UP000075886 UP000076408 UP000075920 UP000075881 UP000075884 UP000030765 UP000094527 UP000198287 UP000075900 UP000069272 UP000076858 UP000000305 UP000054359 UP000053240 UP000007266
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4JMK8
H9JNZ8
A0A2H1VSH3
A0A212EWP5
A0A194PTK0
A0A1Y1LM47
+ More
A0A0T6AZF4 A0A2J7QNA9 A0A1Y1LH82 U4UID2 A0A2P8XSC0 E0VA31 A0A195FXV0 A0A195C0Y5 A0A0M8ZME8 F4WJS3 A0A195BBC1 A0A154P9R3 A0A158NS50 A0A195DE68 E9IFP0 A0A151WH11 A0A3L8DE59 A0A0L7RD67 A0A087ZNK0 E2B8V5 A0A026VZC8 A0A0J7LAH5 A0A2W1BA10 A0A067QY70 A0A1B6E7D8 A0A0C9R3L5 A0A0C9QDM1 T1HSD4 A0A1J1I7R4 A0A023EY15 A0A1L8DJ79 A0A1B6LDD1 A0A1B6HYL7 A0A1Q3FNA6 A0A1Q3FNB6 B0W4J7 Q17K38 A0A1S4EZV1 A0A182G3H7 A0A2M4CFV6 W5JUE7 A0A182LZ69 A0A182IXN2 A0A1S4GY38 Q7Q4G9 A0A182PLS2 A0A182UHU4 A0A182IC57 A0A182VDF7 A0A2C9GNI8 A0A182Q9Z8 A0A182YDT6 A0A182W6F0 A0A182K1H6 A0A3F2Z0M9 A0A182NU05 A0A2R7W2B1 A0A084WH64 A0A1D2NFU2 A0A226D097 A0A182RDS2 A0A182F3G5 A0A0N8D0C4 A0A0P5DYT9 A0A0N8A3N1 A0A0P4ZSJ2 A0A0P5TQ10 A0A0P5P992 A0A0P5YX33 E9G0L5 A0A0N8D187 A0A0N7ZLN1 A0A0P5CC42 A0A0P5TEE5 T1IPD0 A0A0P6I067 A0A2L2YKQ9 A0A087V0C8 A0A194RHY2 A0A0P5KHA2 A0A139WG98 A0A0P5QD11 A0A0P6BES3 T1DCA4 A0A0P5I8A5 A0A0P5WWX7 A0A0P5A7C5 A0A0P5I988 A0A0P5I4W4 A0A0P5QR85 A0A0P6F0X6
A0A0T6AZF4 A0A2J7QNA9 A0A1Y1LH82 U4UID2 A0A2P8XSC0 E0VA31 A0A195FXV0 A0A195C0Y5 A0A0M8ZME8 F4WJS3 A0A195BBC1 A0A154P9R3 A0A158NS50 A0A195DE68 E9IFP0 A0A151WH11 A0A3L8DE59 A0A0L7RD67 A0A087ZNK0 E2B8V5 A0A026VZC8 A0A0J7LAH5 A0A2W1BA10 A0A067QY70 A0A1B6E7D8 A0A0C9R3L5 A0A0C9QDM1 T1HSD4 A0A1J1I7R4 A0A023EY15 A0A1L8DJ79 A0A1B6LDD1 A0A1B6HYL7 A0A1Q3FNA6 A0A1Q3FNB6 B0W4J7 Q17K38 A0A1S4EZV1 A0A182G3H7 A0A2M4CFV6 W5JUE7 A0A182LZ69 A0A182IXN2 A0A1S4GY38 Q7Q4G9 A0A182PLS2 A0A182UHU4 A0A182IC57 A0A182VDF7 A0A2C9GNI8 A0A182Q9Z8 A0A182YDT6 A0A182W6F0 A0A182K1H6 A0A3F2Z0M9 A0A182NU05 A0A2R7W2B1 A0A084WH64 A0A1D2NFU2 A0A226D097 A0A182RDS2 A0A182F3G5 A0A0N8D0C4 A0A0P5DYT9 A0A0N8A3N1 A0A0P4ZSJ2 A0A0P5TQ10 A0A0P5P992 A0A0P5YX33 E9G0L5 A0A0N8D187 A0A0N7ZLN1 A0A0P5CC42 A0A0P5TEE5 T1IPD0 A0A0P6I067 A0A2L2YKQ9 A0A087V0C8 A0A194RHY2 A0A0P5KHA2 A0A139WG98 A0A0P5QD11 A0A0P6BES3 T1DCA4 A0A0P5I8A5 A0A0P5WWX7 A0A0P5A7C5 A0A0P5I988 A0A0P5I4W4 A0A0P5QR85 A0A0P6F0X6
PDB
6MOL
E-value=1.05294e-37,
Score=394
Ontologies
GO
Topology
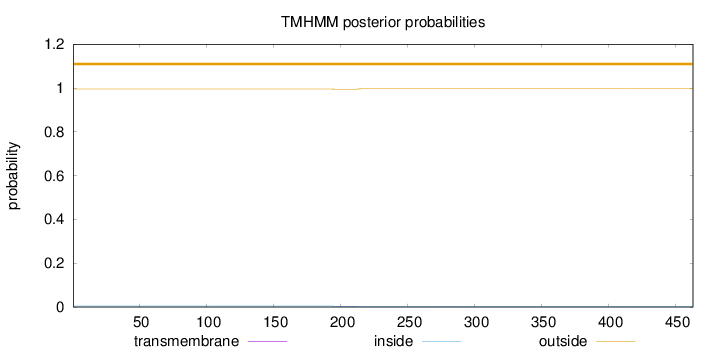
Length:
463
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0531100000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00467
outside
1 - 463
Population Genetic Test Statistics
Pi
244.16981
Theta
175.445934
Tajima's D
1.23599
CLR
0.31665
CSRT
0.715764211789411
Interpretation
Uncertain
