Gene
KWMTBOMO14014
Pre Gene Modal
BGIBMGA011253
Annotation
PREDICTED:_26S_proteasome_complex_subunit_DSS1_[Plutella_xylostella]
Full name
Actin-related protein 8
Location in the cell
Nuclear Reliability : 2.236
Sequence
CDS
ATGGCTGACAAACAGAAAGTAGACCTTGGTTTGCTAGAAGAGGATGACGAGTTCGAAGAATTTCCCGCGGAAAATTGGGGTACAGAAGACGCTGATGATGAAGATGTATCGGTTTGGGAAGACAACTGGGAAGACGACGTCATCCAAGATGATTTTAATCAGCAGTTGAGGCAACAACTGGAAAAACTGAAGGATCAGAAATCTTAA
Protein
MADKQKVDLGLLEEDDEFEEFPAENWGTEDADDEDVSVWEDNWEDDVIQDDFNQQLRQQLEKLKDQKS
Summary
Description
Plays an important role in the functional organization of mitotic chromosomes. Exhibits low basal ATPase activity, and unable to polymerize.
Proposed core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair.
Proposed core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair.
Subunit
Component of the chromatin remodeling Ino80 complex. Exists as monomers and dimers, but the dimer is most probably the biologically relevant form required for stable interactions with histones that exploits the twofold symmetry of the nucleosome core.
Similarity
Belongs to the actin family. ARP8 subfamily.
Uniprot
H9JNZ9
A0A3S2PHG7
A0A2A4JNI9
A0A2H1VQ76
A0A2W1B5R8
A0A194PS84
+ More
A0A194RJC4 A0A212EWN6 S4PH22 N6ST48 D6WHE9 A0A1W4XHR7 A0A067RHA1 A0A1Q3FE76 A0A182WV10 Q7PS61 A0A182I8N5 B0WT20 A0A2J7PWT5 A0A1S3DBK9 T1DGY9 A0A2T7PQM3 Q17J99 A0A023ED46 A0A182IWT9 A0A182K2Q0 A0A182P2N7 A0A182XX42 A0A182QMA5 T1E8X6 A0A2M3ZAV8 A0A2M4ANF6 A0A2M4C5V9 W5JB26 A0A182F9A7 A0A182RVY7 A0A182W153 A0A182NFY8 A0A2R7VWK9 A0A1B0DLB9 A0A0C9RRF2 A0A1E1WYS7 A0A1L8DSL2 A0A293M336 A0A0K8TPG4 V5I4Z0 B7QKW9 A0A131Y7W2 A0A131YER4 L7M251 A0A131XKW3 A0A087U614 A0A1B0CFG2 A0A3B5AZD4 E9HAR1 A0A2G8L2A0 F7FM90 A0A3M6TLK3 A0A0P6ACR8 A0A0P6GWZ3 V8NWK1 E2RK10 A0A3Q3D759 A0A3Q3EG79 A0A060XIN4 A0A1W7REI8 T1E4M4 A0A0F7Z5M7 A0A0B8RPJ2 U3ERI8 F6NMA4 A0A151MQL7 G1KQ62 A0A1A6GF70 I3MHG4 A0A1U7R086 A0A286XPK7 D3ZHW9 Q3TV35 A0A2K6EC95 A0A2K6MCL8 G1U1K1 G1LV61 G3S3V4 G1S1L9 A0A341D6W6 A0A2K5ZAR8 F2Z5F4 A0A0D9RKP1 G3T9T5 A0A2Y9E2X8 A0A2U3V9X3 A0A2I3MP30 A0A2K5EE89 A0A2Y9T2J4 A0A2K6RAT9 A0A2U3XY50 A0A337S1G4 A0A2K5KTJ0 A0A3P4RW88
A0A194RJC4 A0A212EWN6 S4PH22 N6ST48 D6WHE9 A0A1W4XHR7 A0A067RHA1 A0A1Q3FE76 A0A182WV10 Q7PS61 A0A182I8N5 B0WT20 A0A2J7PWT5 A0A1S3DBK9 T1DGY9 A0A2T7PQM3 Q17J99 A0A023ED46 A0A182IWT9 A0A182K2Q0 A0A182P2N7 A0A182XX42 A0A182QMA5 T1E8X6 A0A2M3ZAV8 A0A2M4ANF6 A0A2M4C5V9 W5JB26 A0A182F9A7 A0A182RVY7 A0A182W153 A0A182NFY8 A0A2R7VWK9 A0A1B0DLB9 A0A0C9RRF2 A0A1E1WYS7 A0A1L8DSL2 A0A293M336 A0A0K8TPG4 V5I4Z0 B7QKW9 A0A131Y7W2 A0A131YER4 L7M251 A0A131XKW3 A0A087U614 A0A1B0CFG2 A0A3B5AZD4 E9HAR1 A0A2G8L2A0 F7FM90 A0A3M6TLK3 A0A0P6ACR8 A0A0P6GWZ3 V8NWK1 E2RK10 A0A3Q3D759 A0A3Q3EG79 A0A060XIN4 A0A1W7REI8 T1E4M4 A0A0F7Z5M7 A0A0B8RPJ2 U3ERI8 F6NMA4 A0A151MQL7 G1KQ62 A0A1A6GF70 I3MHG4 A0A1U7R086 A0A286XPK7 D3ZHW9 Q3TV35 A0A2K6EC95 A0A2K6MCL8 G1U1K1 G1LV61 G3S3V4 G1S1L9 A0A341D6W6 A0A2K5ZAR8 F2Z5F4 A0A0D9RKP1 G3T9T5 A0A2Y9E2X8 A0A2U3V9X3 A0A2I3MP30 A0A2K5EE89 A0A2Y9T2J4 A0A2K6RAT9 A0A2U3XY50 A0A337S1G4 A0A2K5KTJ0 A0A3P4RW88
Pubmed
19121390
28756777
26354079
22118469
23622113
23537049
+ More
18362917 19820115 24845553 12364791 14747013 17210077 24330624 17510324 24945155 26483478 25244985 20920257 23761445 26131772 28503490 26369729 25765539 26830274 25576852 28049606 21292972 29023486 18464734 30382153 24297900 16341006 24755649 26358130 23758969 25476704 23915248 23594743 22293439 21993624 15057822 15632090 29398115 10349636 11042159 11076861 11217851 12466851 12040188 16141073 20010809 25362486 17975172
18362917 19820115 24845553 12364791 14747013 17210077 24330624 17510324 24945155 26483478 25244985 20920257 23761445 26131772 28503490 26369729 25765539 26830274 25576852 28049606 21292972 29023486 18464734 30382153 24297900 16341006 24755649 26358130 23758969 25476704 23915248 23594743 22293439 21993624 15057822 15632090 29398115 10349636 11042159 11076861 11217851 12466851 12040188 16141073 20010809 25362486 17975172
EMBL
BABH01009707
RSAL01000031
RVE51587.1
NWSH01000978
PCG73288.1
ODYU01003796
+ More
SOQ43001.1 KZ150299 PZC71542.1 KQ459593 KPI96301.1 KQ460152 KPJ17440.1 AGBW02011924 OWR45902.1 GAIX01005960 JAA86600.1 APGK01057313 KB741280 KB631604 ENN70869.1 ERL84587.1 KQ971321 EFA00655.1 KK852470 KDR23221.1 GFDL01009189 JAV25856.1 AAAB01008844 EAA06113.4 APCN01003090 DS232079 EDS34157.1 NEVH01020869 PNF20791.1 GALA01000002 JAA94850.1 PZQS01000002 PVD35724.1 CH477234 EAT46707.1 JXUM01095178 GAPW01006842 GAPW01006841 KQ564177 JAC06756.1 KXJ72617.1 AXCN02000343 GAMD01002373 JAA99217.1 GGFM01004859 MBW25610.1 GGFK01008966 MBW42287.1 GGFJ01011400 MBW60541.1 ADMH02001636 GGFL01004441 ETN61662.1 MBW68619.1 KK854050 PTY10305.1 AJVK01071098 GBZX01002944 JAG89796.1 GFAC01007023 JAT92165.1 GFDF01004693 JAV09391.1 GFWV01010425 MAA35154.1 GDAI01001321 JAI16282.1 GANP01000810 JAB83658.1 ABJB010256305 DS962443 EEC19491.1 GEFM01001785 JAP74011.1 GEDV01010694 JAP77863.1 GACK01006628 JAA58406.1 GEFH01001781 JAP66800.1 KK118364 KFM72803.1 AJWK01010098 GL732613 EFX71170.1 MRZV01000251 PIK54362.1 RCHS01003372 RMX42317.1 GDIP01031235 JAM72480.1 GDIQ01028243 JAN66494.1 AZIM01001678 ETE66033.1 AAEX03009319 FR905461 CDQ79478.1 GDAY02001895 JAV49537.1 GAAZ01001930 JAA96013.1 GBEX01002654 JAI11906.1 GBSH01002128 GBSH01002117 JAG66909.1 GAEP01001228 GBEW01000758 JAB53593.1 JAI09607.1 CU634012 AKHW03005461 KYO26690.1 LZPO01097125 OBS64876.1 AGTP01068139 AAKN02016906 AAKN02016907 AAKN02016908 AAKN02016909 AABR07072979 CH473959 EDM15031.1 AK160432 AK161690 AK167914 CH466533 BAE35785.1 BAE36534.1 BAE39921.1 EDL13957.1 AAGW02035468 ACTA01050967 ACTA01058967 CABD030053439 CABD030053440 CABD030053441 ADFV01001003 AEMK02000070 AQIB01018663 AHZZ02023056 AANG04003224 CYRY02043418 VCX37798.1
SOQ43001.1 KZ150299 PZC71542.1 KQ459593 KPI96301.1 KQ460152 KPJ17440.1 AGBW02011924 OWR45902.1 GAIX01005960 JAA86600.1 APGK01057313 KB741280 KB631604 ENN70869.1 ERL84587.1 KQ971321 EFA00655.1 KK852470 KDR23221.1 GFDL01009189 JAV25856.1 AAAB01008844 EAA06113.4 APCN01003090 DS232079 EDS34157.1 NEVH01020869 PNF20791.1 GALA01000002 JAA94850.1 PZQS01000002 PVD35724.1 CH477234 EAT46707.1 JXUM01095178 GAPW01006842 GAPW01006841 KQ564177 JAC06756.1 KXJ72617.1 AXCN02000343 GAMD01002373 JAA99217.1 GGFM01004859 MBW25610.1 GGFK01008966 MBW42287.1 GGFJ01011400 MBW60541.1 ADMH02001636 GGFL01004441 ETN61662.1 MBW68619.1 KK854050 PTY10305.1 AJVK01071098 GBZX01002944 JAG89796.1 GFAC01007023 JAT92165.1 GFDF01004693 JAV09391.1 GFWV01010425 MAA35154.1 GDAI01001321 JAI16282.1 GANP01000810 JAB83658.1 ABJB010256305 DS962443 EEC19491.1 GEFM01001785 JAP74011.1 GEDV01010694 JAP77863.1 GACK01006628 JAA58406.1 GEFH01001781 JAP66800.1 KK118364 KFM72803.1 AJWK01010098 GL732613 EFX71170.1 MRZV01000251 PIK54362.1 RCHS01003372 RMX42317.1 GDIP01031235 JAM72480.1 GDIQ01028243 JAN66494.1 AZIM01001678 ETE66033.1 AAEX03009319 FR905461 CDQ79478.1 GDAY02001895 JAV49537.1 GAAZ01001930 JAA96013.1 GBEX01002654 JAI11906.1 GBSH01002128 GBSH01002117 JAG66909.1 GAEP01001228 GBEW01000758 JAB53593.1 JAI09607.1 CU634012 AKHW03005461 KYO26690.1 LZPO01097125 OBS64876.1 AGTP01068139 AAKN02016906 AAKN02016907 AAKN02016908 AAKN02016909 AABR07072979 CH473959 EDM15031.1 AK160432 AK161690 AK167914 CH466533 BAE35785.1 BAE36534.1 BAE39921.1 EDL13957.1 AAGW02035468 ACTA01050967 ACTA01058967 CABD030053439 CABD030053440 CABD030053441 ADFV01001003 AEMK02000070 AQIB01018663 AHZZ02023056 AANG04003224 CYRY02043418 VCX37798.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000019118 UP000030742 UP000007266 UP000192223 UP000027135 UP000076407 UP000007062 UP000075840 UP000002320 UP000235965 UP000079169 UP000245119 UP000008820 UP000069940 UP000249989 UP000075880 UP000075881 UP000075885 UP000076408 UP000075886 UP000000673 UP000069272 UP000075900 UP000075920 UP000075884 UP000092462 UP000001555 UP000054359 UP000092461 UP000261400 UP000000305 UP000230750 UP000002279 UP000275408 UP000002254 UP000264820 UP000261660 UP000193380 UP000000437 UP000050525 UP000001646 UP000092124 UP000005215 UP000189706 UP000005447 UP000002494 UP000233120 UP000233180 UP000001811 UP000008912 UP000001519 UP000001073 UP000252040 UP000233140 UP000008227 UP000029965 UP000007646 UP000248480 UP000245320 UP000028761 UP000233020 UP000248484 UP000233200 UP000245341 UP000011712 UP000233060
UP000019118 UP000030742 UP000007266 UP000192223 UP000027135 UP000076407 UP000007062 UP000075840 UP000002320 UP000235965 UP000079169 UP000245119 UP000008820 UP000069940 UP000249989 UP000075880 UP000075881 UP000075885 UP000076408 UP000075886 UP000000673 UP000069272 UP000075900 UP000075920 UP000075884 UP000092462 UP000001555 UP000054359 UP000092461 UP000261400 UP000000305 UP000230750 UP000002279 UP000275408 UP000002254 UP000264820 UP000261660 UP000193380 UP000000437 UP000050525 UP000001646 UP000092124 UP000005215 UP000189706 UP000005447 UP000002494 UP000233120 UP000233180 UP000001811 UP000008912 UP000001519 UP000001073 UP000252040 UP000233140 UP000008227 UP000029965 UP000007646 UP000248480 UP000245320 UP000028761 UP000233020 UP000248484 UP000233200 UP000245341 UP000011712 UP000233060
ProteinModelPortal
H9JNZ9
A0A3S2PHG7
A0A2A4JNI9
A0A2H1VQ76
A0A2W1B5R8
A0A194PS84
+ More
A0A194RJC4 A0A212EWN6 S4PH22 N6ST48 D6WHE9 A0A1W4XHR7 A0A067RHA1 A0A1Q3FE76 A0A182WV10 Q7PS61 A0A182I8N5 B0WT20 A0A2J7PWT5 A0A1S3DBK9 T1DGY9 A0A2T7PQM3 Q17J99 A0A023ED46 A0A182IWT9 A0A182K2Q0 A0A182P2N7 A0A182XX42 A0A182QMA5 T1E8X6 A0A2M3ZAV8 A0A2M4ANF6 A0A2M4C5V9 W5JB26 A0A182F9A7 A0A182RVY7 A0A182W153 A0A182NFY8 A0A2R7VWK9 A0A1B0DLB9 A0A0C9RRF2 A0A1E1WYS7 A0A1L8DSL2 A0A293M336 A0A0K8TPG4 V5I4Z0 B7QKW9 A0A131Y7W2 A0A131YER4 L7M251 A0A131XKW3 A0A087U614 A0A1B0CFG2 A0A3B5AZD4 E9HAR1 A0A2G8L2A0 F7FM90 A0A3M6TLK3 A0A0P6ACR8 A0A0P6GWZ3 V8NWK1 E2RK10 A0A3Q3D759 A0A3Q3EG79 A0A060XIN4 A0A1W7REI8 T1E4M4 A0A0F7Z5M7 A0A0B8RPJ2 U3ERI8 F6NMA4 A0A151MQL7 G1KQ62 A0A1A6GF70 I3MHG4 A0A1U7R086 A0A286XPK7 D3ZHW9 Q3TV35 A0A2K6EC95 A0A2K6MCL8 G1U1K1 G1LV61 G3S3V4 G1S1L9 A0A341D6W6 A0A2K5ZAR8 F2Z5F4 A0A0D9RKP1 G3T9T5 A0A2Y9E2X8 A0A2U3V9X3 A0A2I3MP30 A0A2K5EE89 A0A2Y9T2J4 A0A2K6RAT9 A0A2U3XY50 A0A337S1G4 A0A2K5KTJ0 A0A3P4RW88
A0A194RJC4 A0A212EWN6 S4PH22 N6ST48 D6WHE9 A0A1W4XHR7 A0A067RHA1 A0A1Q3FE76 A0A182WV10 Q7PS61 A0A182I8N5 B0WT20 A0A2J7PWT5 A0A1S3DBK9 T1DGY9 A0A2T7PQM3 Q17J99 A0A023ED46 A0A182IWT9 A0A182K2Q0 A0A182P2N7 A0A182XX42 A0A182QMA5 T1E8X6 A0A2M3ZAV8 A0A2M4ANF6 A0A2M4C5V9 W5JB26 A0A182F9A7 A0A182RVY7 A0A182W153 A0A182NFY8 A0A2R7VWK9 A0A1B0DLB9 A0A0C9RRF2 A0A1E1WYS7 A0A1L8DSL2 A0A293M336 A0A0K8TPG4 V5I4Z0 B7QKW9 A0A131Y7W2 A0A131YER4 L7M251 A0A131XKW3 A0A087U614 A0A1B0CFG2 A0A3B5AZD4 E9HAR1 A0A2G8L2A0 F7FM90 A0A3M6TLK3 A0A0P6ACR8 A0A0P6GWZ3 V8NWK1 E2RK10 A0A3Q3D759 A0A3Q3EG79 A0A060XIN4 A0A1W7REI8 T1E4M4 A0A0F7Z5M7 A0A0B8RPJ2 U3ERI8 F6NMA4 A0A151MQL7 G1KQ62 A0A1A6GF70 I3MHG4 A0A1U7R086 A0A286XPK7 D3ZHW9 Q3TV35 A0A2K6EC95 A0A2K6MCL8 G1U1K1 G1LV61 G3S3V4 G1S1L9 A0A341D6W6 A0A2K5ZAR8 F2Z5F4 A0A0D9RKP1 G3T9T5 A0A2Y9E2X8 A0A2U3V9X3 A0A2I3MP30 A0A2K5EE89 A0A2Y9T2J4 A0A2K6RAT9 A0A2U3XY50 A0A337S1G4 A0A2K5KTJ0 A0A3P4RW88
PDB
6MSK
E-value=0.01871,
Score=82
Ontologies
KEGG
PATHWAY
GO
PANTHER
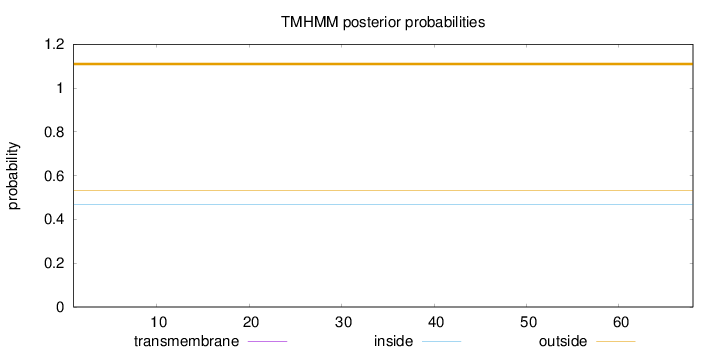
Topology
Subcellular location
Nucleus
Length:
68
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.46735
outside
1 - 68
Population Genetic Test Statistics
Pi
252.941
Theta
197.982363
Tajima's D
0.870725
CLR
0.002861
CSRT
0.628718564071796
Interpretation
Uncertain
