Pre Gene Modal
BGIBMGA004873
Annotation
PREDICTED:_polyribonucleotide_nucleotidyltransferase_1?_mitochondrial-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.983
Sequence
CDS
ATGTCAAAGCCATTCGATCCTTCTCAGGGCATAGAAGACTACATGGGCGATATGGACTTCAAAGTGGCCGGCACGAAGAAGGGCATCACGGCGCTGCAGGCGGACATCAAGCTGGCCGGCCTGCCGCTCCGCGTCGTCATGGAGGCCGTGCAGCGGGCCTGCGACGCCAATGCCAAGATCATCGACATTATGAACCAATGCCTCGACGCACCCAGACAAGGATTAAAAGAGAACATGCCAGTGATTGAAGAGATCGAAGTGGAAGCTCACAAGCGGCCCAAGCTCCTCGGGCTCGGCGGCTCCAACTTGAAGAAGCTCTACGTTGAGACCGGCGTGCAGGTAAGACATCTAGACTATTAA
Protein
MSKPFDPSQGIEDYMGDMDFKVAGTKKGITALQADIKLAGLPLRVVMEAVQRACDANAKIIDIMNQCLDAPRQGLKENMPVIEEIEVEAHKRPKLLGLGGSNLKKLYVETGVQVRHLDY
Summary
Uniprot
H9J5T3
A0A2W1BBA6
A0A2H1WL21
A0A2A4JVN5
A0A2A4JVY3
A0A212FPY4
+ More
A0A194RHJ5 A0A2J7PKR5 D6WRI0 N6U217 A0A2J7PKS3 A0A067QS60 A0A0T6B4Y5 A0A2J7PKR7 A0A1B6LA11 A0A336M3M1 A0A1Y1MHJ5 A0A2J7PKS5 A0A1Y1MFU5 A0A226ENF1 A0A0P5B9V6 A0A1B6M4S1 E9GI57 A0A0K8T125 A0A0A9VR83 A0A1D5QY48 A0A0P5UUU7 A0A1D1V9G5 A0A0N8CDM2 A0A1Z5LEV5 A0A026WFK7 A0A1B6HG47 A0A023GLF8 A0A293N476 A0A0P5PC46 A0A3Q0J0A3 A0A232ELF4 K7IR75 A0A1B6D1K8 A0A182IR73 A0A1Y1MEJ8 A0A0P4ZH66 A0A0P5EJ68 G3MKA0 A0A0P5Z0E1 A0A0P6BFE1 A0A1B6G5H2 A0A2R7W850 A0A0N7ZU02 A0A0P5N2G4 A0A0P5IKB5 A0A084VB39 A0A3Q3K0C5 A0A2R5L4P2 A0A3Q3KCV4 A0A3Q3KBI2 A0A1J1J463 A0A3B5B9M7 A0A3L8DLL9 A0A3Q2CRM7 A0A3B5B9I5 A0A131XRJ5 A0A3P9IDC9 A0A131YY19 A0A131Y0A2 V5HZY0 A0A224Z6D3 A0A3Q0RL98 A0A146VC09 A0A3P9JM96 A0A3B5MDH9 G9KHK7 A0A146MPN6 H2LHG8 A0A146MQ27 A0A3Q4AXU4 A0A3Q3H163 A0A146MSI8 A0A0S7GJB5 A0A3Q4HHA6 A0A1B0GPF7 A0A146MRP7 A0A3Q3H145 A0A3Q4MQ15 A0A3P9N4C9 A0A3B5Q4B3 M4AR70 A0A2I4AYL3 A0A146VD49 A0A3Q2P1Q6 A0A3B3Y6A4 A0A3Q2SXZ3 A0A3B3VEV8 A0A3B3Y6B1 A0A3P8R3V9 A0A3Q2U4N1
A0A194RHJ5 A0A2J7PKR5 D6WRI0 N6U217 A0A2J7PKS3 A0A067QS60 A0A0T6B4Y5 A0A2J7PKR7 A0A1B6LA11 A0A336M3M1 A0A1Y1MHJ5 A0A2J7PKS5 A0A1Y1MFU5 A0A226ENF1 A0A0P5B9V6 A0A1B6M4S1 E9GI57 A0A0K8T125 A0A0A9VR83 A0A1D5QY48 A0A0P5UUU7 A0A1D1V9G5 A0A0N8CDM2 A0A1Z5LEV5 A0A026WFK7 A0A1B6HG47 A0A023GLF8 A0A293N476 A0A0P5PC46 A0A3Q0J0A3 A0A232ELF4 K7IR75 A0A1B6D1K8 A0A182IR73 A0A1Y1MEJ8 A0A0P4ZH66 A0A0P5EJ68 G3MKA0 A0A0P5Z0E1 A0A0P6BFE1 A0A1B6G5H2 A0A2R7W850 A0A0N7ZU02 A0A0P5N2G4 A0A0P5IKB5 A0A084VB39 A0A3Q3K0C5 A0A2R5L4P2 A0A3Q3KCV4 A0A3Q3KBI2 A0A1J1J463 A0A3B5B9M7 A0A3L8DLL9 A0A3Q2CRM7 A0A3B5B9I5 A0A131XRJ5 A0A3P9IDC9 A0A131YY19 A0A131Y0A2 V5HZY0 A0A224Z6D3 A0A3Q0RL98 A0A146VC09 A0A3P9JM96 A0A3B5MDH9 G9KHK7 A0A146MPN6 H2LHG8 A0A146MQ27 A0A3Q4AXU4 A0A3Q3H163 A0A146MSI8 A0A0S7GJB5 A0A3Q4HHA6 A0A1B0GPF7 A0A146MRP7 A0A3Q3H145 A0A3Q4MQ15 A0A3P9N4C9 A0A3B5Q4B3 M4AR70 A0A2I4AYL3 A0A146VD49 A0A3Q2P1Q6 A0A3B3Y6A4 A0A3Q2SXZ3 A0A3B3VEV8 A0A3B3Y6B1 A0A3P8R3V9 A0A3Q2U4N1
Pubmed
EMBL
BABH01025198
BABH01025199
BABH01025200
BABH01025201
KZ150371
PZC71144.1
+ More
ODYU01009033 SOQ53144.1 NWSH01000577 PCG75543.1 PCG75542.1 AGBW02000220 OWR55807.1 KQ460205 KPJ16795.1 NEVH01024531 PNF16934.1 KQ971351 EFA06447.1 APGK01045628 KB741037 KB632319 ENN74666.1 ERL92290.1 PNF16930.1 KK853697 KDQ96253.1 LJIG01009785 KRT82386.1 PNF16932.1 GEBQ01019427 JAT20550.1 UFQT01000509 SSX24854.1 GEZM01036370 JAV82757.1 PNF16933.1 GEZM01036371 JAV82756.1 LNIX01000002 OXA59195.1 GDIP01187468 JAJ35934.1 GEBQ01009055 JAT30922.1 GL732546 EFX80827.1 GBRD01006584 JAG59237.1 GBHO01044672 GDHC01009054 JAF98931.1 JAQ09575.1 JSUE03010920 JSUE03010921 JSUE03010922 GDIP01125037 JAL78677.1 BDGG01000004 GAU98331.1 GDIP01142071 JAL61643.1 GFJQ02001037 JAW05933.1 KK107238 EZA54837.1 GECU01034053 JAS73653.1 GBBM01000661 JAC34757.1 GFWV01023145 MAA47872.1 GDIQ01132870 JAL18856.1 NNAY01003564 OXU19193.1 GEDC01017707 JAS19591.1 GEZM01036372 JAV82755.1 GDIP01217174 JAJ06228.1 GDIP01159182 JAJ64220.1 JO842301 AEO33918.1 GDIP01050745 LRGB01000704 JAM52970.1 KZS16503.1 GDIP01015966 JAM87749.1 GECZ01012206 JAS57563.1 KK854443 PTY15892.1 GDIP01212721 JAJ10681.1 GDIQ01148270 JAL03456.1 GDIQ01220148 JAK31577.1 ATLV01005272 ATLV01005273 KE524276 KFB35183.1 GGLE01000358 MBY04484.1 CVRI01000070 CRL07195.1 QOIP01000007 RLU20809.1 GEFH01000410 JAP68171.1 GEDV01005182 JAP83375.1 GEFM01002883 JAP72913.1 GANP01004130 JAB80338.1 GFPF01010704 MAA21850.1 GCES01071417 JAR14906.1 JP015784 AES04382.1 GCES01164674 JAQ21648.1 GCES01164673 JAQ21649.1 GCES01164672 JAQ21650.1 GBYX01453275 JAO28240.1 AJVK01033652 GCES01164675 JAQ21647.1 GCES01071416 JAR14907.1
ODYU01009033 SOQ53144.1 NWSH01000577 PCG75543.1 PCG75542.1 AGBW02000220 OWR55807.1 KQ460205 KPJ16795.1 NEVH01024531 PNF16934.1 KQ971351 EFA06447.1 APGK01045628 KB741037 KB632319 ENN74666.1 ERL92290.1 PNF16930.1 KK853697 KDQ96253.1 LJIG01009785 KRT82386.1 PNF16932.1 GEBQ01019427 JAT20550.1 UFQT01000509 SSX24854.1 GEZM01036370 JAV82757.1 PNF16933.1 GEZM01036371 JAV82756.1 LNIX01000002 OXA59195.1 GDIP01187468 JAJ35934.1 GEBQ01009055 JAT30922.1 GL732546 EFX80827.1 GBRD01006584 JAG59237.1 GBHO01044672 GDHC01009054 JAF98931.1 JAQ09575.1 JSUE03010920 JSUE03010921 JSUE03010922 GDIP01125037 JAL78677.1 BDGG01000004 GAU98331.1 GDIP01142071 JAL61643.1 GFJQ02001037 JAW05933.1 KK107238 EZA54837.1 GECU01034053 JAS73653.1 GBBM01000661 JAC34757.1 GFWV01023145 MAA47872.1 GDIQ01132870 JAL18856.1 NNAY01003564 OXU19193.1 GEDC01017707 JAS19591.1 GEZM01036372 JAV82755.1 GDIP01217174 JAJ06228.1 GDIP01159182 JAJ64220.1 JO842301 AEO33918.1 GDIP01050745 LRGB01000704 JAM52970.1 KZS16503.1 GDIP01015966 JAM87749.1 GECZ01012206 JAS57563.1 KK854443 PTY15892.1 GDIP01212721 JAJ10681.1 GDIQ01148270 JAL03456.1 GDIQ01220148 JAK31577.1 ATLV01005272 ATLV01005273 KE524276 KFB35183.1 GGLE01000358 MBY04484.1 CVRI01000070 CRL07195.1 QOIP01000007 RLU20809.1 GEFH01000410 JAP68171.1 GEDV01005182 JAP83375.1 GEFM01002883 JAP72913.1 GANP01004130 JAB80338.1 GFPF01010704 MAA21850.1 GCES01071417 JAR14906.1 JP015784 AES04382.1 GCES01164674 JAQ21648.1 GCES01164673 JAQ21649.1 GCES01164672 JAQ21650.1 GBYX01453275 JAO28240.1 AJVK01033652 GCES01164675 JAQ21647.1 GCES01071416 JAR14907.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000235965
UP000007266
+ More
UP000019118 UP000030742 UP000027135 UP000198287 UP000000305 UP000006718 UP000186922 UP000053097 UP000079169 UP000215335 UP000002358 UP000075880 UP000076858 UP000030765 UP000261600 UP000183832 UP000261400 UP000279307 UP000265020 UP000265200 UP000261340 UP000261380 UP000001038 UP000261620 UP000261660 UP000261580 UP000092462 UP000242638 UP000002852 UP000192220 UP000265000 UP000261480 UP000261500 UP000265100
UP000019118 UP000030742 UP000027135 UP000198287 UP000000305 UP000006718 UP000186922 UP000053097 UP000079169 UP000215335 UP000002358 UP000075880 UP000076858 UP000030765 UP000261600 UP000183832 UP000261400 UP000279307 UP000265020 UP000265200 UP000261340 UP000261380 UP000001038 UP000261620 UP000261660 UP000261580 UP000092462 UP000242638 UP000002852 UP000192220 UP000265000 UP000261480 UP000261500 UP000265100
Pfam
Interpro
IPR036612
KH_dom_type_1_sf
+ More
IPR027408 PNPase/RNase_PH_dom_sf
IPR012162 PNPase
IPR012340 NA-bd_OB-fold
IPR015848 PNPase_PH_RNA-bd_bac/org-type
IPR001247 ExoRNase_PH_dom1
IPR015847 ExoRNase_PH_dom2
IPR036345 ExoRNase_PH_dom2_sf
IPR003029 S1_domain
IPR020568 Ribosomal_S5_D2-typ_fold
IPR004087 KH_dom
IPR036456 PNPase_PH_RNA-bd_sf
IPR004088 KH_dom_type_1
IPR036621 Anticodon-bd_dom_sf
IPR041715 HisRS-like_core
IPR024435 HisRS-related_dom
IPR022967 S1_dom
IPR027408 PNPase/RNase_PH_dom_sf
IPR012162 PNPase
IPR012340 NA-bd_OB-fold
IPR015848 PNPase_PH_RNA-bd_bac/org-type
IPR001247 ExoRNase_PH_dom1
IPR015847 ExoRNase_PH_dom2
IPR036345 ExoRNase_PH_dom2_sf
IPR003029 S1_domain
IPR020568 Ribosomal_S5_D2-typ_fold
IPR004087 KH_dom
IPR036456 PNPase_PH_RNA-bd_sf
IPR004088 KH_dom_type_1
IPR036621 Anticodon-bd_dom_sf
IPR041715 HisRS-like_core
IPR024435 HisRS-related_dom
IPR022967 S1_dom
SUPFAM
Gene 3D
ProteinModelPortal
H9J5T3
A0A2W1BBA6
A0A2H1WL21
A0A2A4JVN5
A0A2A4JVY3
A0A212FPY4
+ More
A0A194RHJ5 A0A2J7PKR5 D6WRI0 N6U217 A0A2J7PKS3 A0A067QS60 A0A0T6B4Y5 A0A2J7PKR7 A0A1B6LA11 A0A336M3M1 A0A1Y1MHJ5 A0A2J7PKS5 A0A1Y1MFU5 A0A226ENF1 A0A0P5B9V6 A0A1B6M4S1 E9GI57 A0A0K8T125 A0A0A9VR83 A0A1D5QY48 A0A0P5UUU7 A0A1D1V9G5 A0A0N8CDM2 A0A1Z5LEV5 A0A026WFK7 A0A1B6HG47 A0A023GLF8 A0A293N476 A0A0P5PC46 A0A3Q0J0A3 A0A232ELF4 K7IR75 A0A1B6D1K8 A0A182IR73 A0A1Y1MEJ8 A0A0P4ZH66 A0A0P5EJ68 G3MKA0 A0A0P5Z0E1 A0A0P6BFE1 A0A1B6G5H2 A0A2R7W850 A0A0N7ZU02 A0A0P5N2G4 A0A0P5IKB5 A0A084VB39 A0A3Q3K0C5 A0A2R5L4P2 A0A3Q3KCV4 A0A3Q3KBI2 A0A1J1J463 A0A3B5B9M7 A0A3L8DLL9 A0A3Q2CRM7 A0A3B5B9I5 A0A131XRJ5 A0A3P9IDC9 A0A131YY19 A0A131Y0A2 V5HZY0 A0A224Z6D3 A0A3Q0RL98 A0A146VC09 A0A3P9JM96 A0A3B5MDH9 G9KHK7 A0A146MPN6 H2LHG8 A0A146MQ27 A0A3Q4AXU4 A0A3Q3H163 A0A146MSI8 A0A0S7GJB5 A0A3Q4HHA6 A0A1B0GPF7 A0A146MRP7 A0A3Q3H145 A0A3Q4MQ15 A0A3P9N4C9 A0A3B5Q4B3 M4AR70 A0A2I4AYL3 A0A146VD49 A0A3Q2P1Q6 A0A3B3Y6A4 A0A3Q2SXZ3 A0A3B3VEV8 A0A3B3Y6B1 A0A3P8R3V9 A0A3Q2U4N1
A0A194RHJ5 A0A2J7PKR5 D6WRI0 N6U217 A0A2J7PKS3 A0A067QS60 A0A0T6B4Y5 A0A2J7PKR7 A0A1B6LA11 A0A336M3M1 A0A1Y1MHJ5 A0A2J7PKS5 A0A1Y1MFU5 A0A226ENF1 A0A0P5B9V6 A0A1B6M4S1 E9GI57 A0A0K8T125 A0A0A9VR83 A0A1D5QY48 A0A0P5UUU7 A0A1D1V9G5 A0A0N8CDM2 A0A1Z5LEV5 A0A026WFK7 A0A1B6HG47 A0A023GLF8 A0A293N476 A0A0P5PC46 A0A3Q0J0A3 A0A232ELF4 K7IR75 A0A1B6D1K8 A0A182IR73 A0A1Y1MEJ8 A0A0P4ZH66 A0A0P5EJ68 G3MKA0 A0A0P5Z0E1 A0A0P6BFE1 A0A1B6G5H2 A0A2R7W850 A0A0N7ZU02 A0A0P5N2G4 A0A0P5IKB5 A0A084VB39 A0A3Q3K0C5 A0A2R5L4P2 A0A3Q3KCV4 A0A3Q3KBI2 A0A1J1J463 A0A3B5B9M7 A0A3L8DLL9 A0A3Q2CRM7 A0A3B5B9I5 A0A131XRJ5 A0A3P9IDC9 A0A131YY19 A0A131Y0A2 V5HZY0 A0A224Z6D3 A0A3Q0RL98 A0A146VC09 A0A3P9JM96 A0A3B5MDH9 G9KHK7 A0A146MPN6 H2LHG8 A0A146MQ27 A0A3Q4AXU4 A0A3Q3H163 A0A146MSI8 A0A0S7GJB5 A0A3Q4HHA6 A0A1B0GPF7 A0A146MRP7 A0A3Q3H145 A0A3Q4MQ15 A0A3P9N4C9 A0A3B5Q4B3 M4AR70 A0A2I4AYL3 A0A146VD49 A0A3Q2P1Q6 A0A3B3Y6A4 A0A3Q2SXZ3 A0A3B3VEV8 A0A3B3Y6B1 A0A3P8R3V9 A0A3Q2U4N1
PDB
3U1K
E-value=7.94156e-25,
Score=275
Ontologies
GO
PANTHER
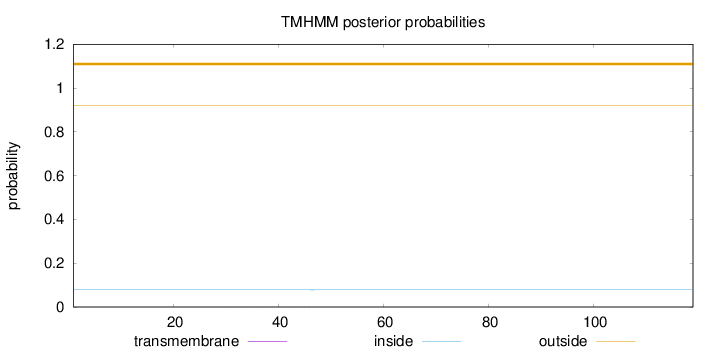
Topology
Length:
119
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01174
Exp number, first 60 AAs:
0.01174
Total prob of N-in:
0.07826
outside
1 - 119
Population Genetic Test Statistics
Pi
324.262413
Theta
243.65672
Tajima's D
0.923642
CLR
1047.677727
CSRT
0.638618069096545
Interpretation
Uncertain
