Gene
KWMTBOMO14008
Pre Gene Modal
BGIBMGA011258
Annotation
PREDICTED:_metabotropic_glutamate_receptor_5-like_[Bombyx_mori]
Full name
Metabotropic glutamate receptor 1
Location in the cell
Extracellular Reliability : 1.069 Mitochondrial Reliability : 1.421
Sequence
CDS
ATGTTGGAGGTGTTAGCGGTGCTGGTGACTGCGCTGCTGGTTCCCGCGGACGCGGAGCGGGCCGTGGTGCGGGGCGCAGTGCGACTGGGAGCCCTGTTCGCAGTGCACGGAGCGCCCGCGGAGGCCGGCGCTCGCTGCGGGCCCGTGCGGGAGCACTACGGAATCCAGCGCGTGGAGGCAACGCTGCGGGCGTTAGATGCAATCAACGCGGACGGATCGCTCCTGCCGGGACTGCAGCTGGGCGCCGAGCTCCGCGACTCGTGCTGGGCGCCCTCAACCGCGCTCCGAGAGACCATCGAGCTGGTGCGAGACGCCATCGCCCCGGCACGTGCGCGTCAGCACTCGCCAGCAGCCTCCGCCGCTAACTGCACTTCGGTATACGATATGCTTTATGCTCTTATCGGTTTTTATCGATGA
Protein
MLEVLAVLVTALLVPADAERAVVRGAVRLGALFAVHGAPAEAGARCGPVREHYGIQRVEATLRALDAINADGSLLPGLQLGAELRDSCWAPSTALRETIELVRDAIAPARARQHSPAASAANCTSVYDMLYALIGFYR
Summary
Subunit
Homodimer; disulfide-linked. The PPXXF motif binds HOMER1, HOMER2 and HOMER3. Interacts with SIAH1, RYR1, RYR2, ITPR1, SHANK1, SHANK3 and GRASP (By similarity).
Similarity
Belongs to the G-protein coupled receptor 3 family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
Disulfide bond
G-protein coupled receptor
Glycoprotein
Membrane
Phosphoprotein
Receptor
Reference proteome
Signal
Transducer
Transmembrane
Transmembrane helix
Feature
chain Metabotropic glutamate receptor 1
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
H9JP04
A0A2H1VBH7
V5I6D0
A0A087UMR8
A0A2I0U879
D2HKF5
+ More
H9FJ34 X1WE31 A0A2P4T7C1 L5KGK8 A0A1J0FAU4 A0A1L8HJI1 Q1ZZG7 G3WUM8 A0A061IJ43 G1NIV0 A0A2K5U693 A0A2K5YDW9 P97772-3 L5MDQ1 A0A3M0KP77 I7G6G0 A0A0D9RX30 G3PCT4 A0A3Q2GRW9 L9LDV8 S9YAF4 A0A0P7TWJ2 A0A226PQQ4 A0A2P4SZW7 A0A3B3RBI0 D6WA59 F6VEM8 H3A0A3 A0A3B3RBE6 A0A1S3SR73 A0A1S3SR84 A0A1S3SR79
H9FJ34 X1WE31 A0A2P4T7C1 L5KGK8 A0A1J0FAU4 A0A1L8HJI1 Q1ZZG7 G3WUM8 A0A061IJ43 G1NIV0 A0A2K5U693 A0A2K5YDW9 P97772-3 L5MDQ1 A0A3M0KP77 I7G6G0 A0A0D9RX30 G3PCT4 A0A3Q2GRW9 L9LDV8 S9YAF4 A0A0P7TWJ2 A0A226PQQ4 A0A2P4SZW7 A0A3B3RBI0 D6WA59 F6VEM8 H3A0A3 A0A3B3RBE6 A0A1S3SR73 A0A1S3SR84 A0A1S3SR79
Pubmed
EMBL
BABH01009694
ODYU01001660
SOQ38193.1
GALX01008400
JAB60066.1
KK120608
+ More
KFM78657.1 KZ506016 PKU42238.1 GL192950 EFB26564.1 JU330887 AFE74643.1 BX296541 CR392011 FO704565 PPHD01006327 POI32246.1 KB030797 ELK09648.1 KU986886 APC26115.1 CM004468 OCT96228.1 DQ417741 ABD74423.1 AEFK01173063 KE667936 ERE84461.1 AQIA01050144 AQIA01050145 AQIA01050146 AQIA01050147 AQIA01050148 AQIA01050149 AQIA01050150 AQIA01050151 AQIA01050152 AF320126 BC067057 BC079566 U89891 KB101593 ELK36481.1 QRBI01000106 RMC12810.1 AB172892 BAE89954.1 AQIB01119431 AQIB01119432 AQIB01119433 AQIB01119434 AQIB01119435 AQIB01119436 AQIB01119437 AQIB01119438 AQIB01119439 AQIB01119440 KB320384 ELW72894.1 KB016759 EPY84451.1 JARO02014523 KPP58158.1 AWGT02000025 OXB82416.1 PPHD01014481 POI29657.1 KQ971312 EEZ98591.1 AAPN01225187 AAPN01225188 AAPN01225189 AAPN01225190 AAPN01225191 AAPN01225192 AAPN01225193 AAPN01225194 AAPN01225195 AFYH01196982 AFYH01196983 AFYH01196984 AFYH01196985 AFYH01196986 AFYH01196987 AFYH01196988 AFYH01196989 AFYH01196990 AFYH01196991
KFM78657.1 KZ506016 PKU42238.1 GL192950 EFB26564.1 JU330887 AFE74643.1 BX296541 CR392011 FO704565 PPHD01006327 POI32246.1 KB030797 ELK09648.1 KU986886 APC26115.1 CM004468 OCT96228.1 DQ417741 ABD74423.1 AEFK01173063 KE667936 ERE84461.1 AQIA01050144 AQIA01050145 AQIA01050146 AQIA01050147 AQIA01050148 AQIA01050149 AQIA01050150 AQIA01050151 AQIA01050152 AF320126 BC067057 BC079566 U89891 KB101593 ELK36481.1 QRBI01000106 RMC12810.1 AB172892 BAE89954.1 AQIB01119431 AQIB01119432 AQIB01119433 AQIB01119434 AQIB01119435 AQIB01119436 AQIB01119437 AQIB01119438 AQIB01119439 AQIB01119440 KB320384 ELW72894.1 KB016759 EPY84451.1 JARO02014523 KPP58158.1 AWGT02000025 OXB82416.1 PPHD01014481 POI29657.1 KQ971312 EEZ98591.1 AAPN01225187 AAPN01225188 AAPN01225189 AAPN01225190 AAPN01225191 AAPN01225192 AAPN01225193 AAPN01225194 AAPN01225195 AFYH01196982 AFYH01196983 AFYH01196984 AFYH01196985 AFYH01196986 AFYH01196987 AFYH01196988 AFYH01196989 AFYH01196990 AFYH01196991
Proteomes
Interpro
SUPFAM
SSF53822
SSF53822
Gene 3D
ProteinModelPortal
H9JP04
A0A2H1VBH7
V5I6D0
A0A087UMR8
A0A2I0U879
D2HKF5
+ More
H9FJ34 X1WE31 A0A2P4T7C1 L5KGK8 A0A1J0FAU4 A0A1L8HJI1 Q1ZZG7 G3WUM8 A0A061IJ43 G1NIV0 A0A2K5U693 A0A2K5YDW9 P97772-3 L5MDQ1 A0A3M0KP77 I7G6G0 A0A0D9RX30 G3PCT4 A0A3Q2GRW9 L9LDV8 S9YAF4 A0A0P7TWJ2 A0A226PQQ4 A0A2P4SZW7 A0A3B3RBI0 D6WA59 F6VEM8 H3A0A3 A0A3B3RBE6 A0A1S3SR73 A0A1S3SR84 A0A1S3SR79
H9FJ34 X1WE31 A0A2P4T7C1 L5KGK8 A0A1J0FAU4 A0A1L8HJI1 Q1ZZG7 G3WUM8 A0A061IJ43 G1NIV0 A0A2K5U693 A0A2K5YDW9 P97772-3 L5MDQ1 A0A3M0KP77 I7G6G0 A0A0D9RX30 G3PCT4 A0A3Q2GRW9 L9LDV8 S9YAF4 A0A0P7TWJ2 A0A226PQQ4 A0A2P4SZW7 A0A3B3RBI0 D6WA59 F6VEM8 H3A0A3 A0A3B3RBE6 A0A1S3SR73 A0A1S3SR84 A0A1S3SR79
PDB
1ISS
E-value=6.46155e-22,
Score=250
Ontologies
GO
GO:0004930
GO:0016021
GO:0051966
GO:0099530
GO:0008066
GO:0098839
GO:0099583
GO:0007216
GO:0005887
GO:0005886
GO:0000187
GO:0001639
GO:0098685
GO:0043025
GO:0007626
GO:0005634
GO:0071257
GO:0019233
GO:0038038
GO:0099061
GO:0000186
GO:0043197
GO:0042734
GO:0051930
GO:0098872
GO:0014069
GO:0045211
GO:0019722
GO:0043408
GO:0038037
GO:0030425
GO:0043005
GO:0030424
GO:0098978
GO:0042802
GO:0051482
GO:0099055
GO:0030331
GO:0007186
GO:0016020
GO:0004713
GO:0016491
GO:0015930
Topology
Subcellular location
Cell membrane
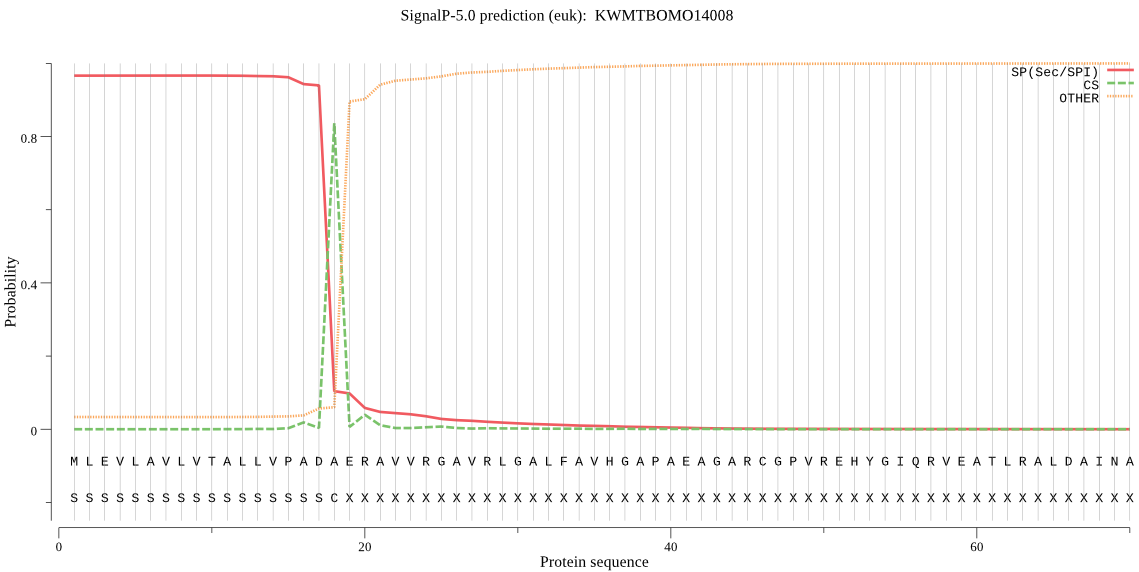
SignalP
Position: 1 - 18,
Likelihood: 0.966121
Length:
138
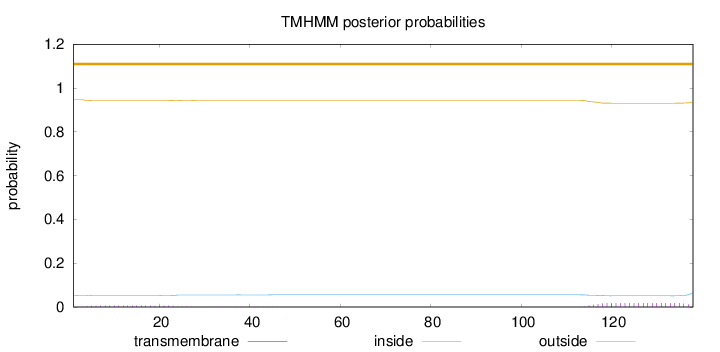
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.52046
Exp number, first 60 AAs:
0.13501
Total prob of N-in:
0.05374
outside
1 - 138
Population Genetic Test Statistics
Pi
314.877895
Theta
209.541321
Tajima's D
1.578818
CLR
135.782516
CSRT
0.801709914504275
Interpretation
Uncertain
