Gene
KWMTBOMO14000 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011344
Annotation
cathepsin_D_precursor_[Bombyx_mori]
Full name
Lysosomal aspartic protease
Location in the cell
Lysosomal Reliability : 1.415 PlasmaMembrane Reliability : 1.634
Sequence
CDS
ATGGGAAAGATATCTTTATTTTTTTTGGCGCTGATCGCCAGCTCCGTAATGGCACTATATAGGGTGCCATTACATCGTATGAAAACTGCGAGAACCCACTTTCATGAGGTTGGCACTGAACTGGAGCTGTTGAGATTAAAATACGATGTGACTGGCCCTTCACCCGAACCATTGTCAAATTATCTTGATGCTCAGTACTACGGAGTGATCAGTATCGGCACGCCGCCGCAGTCGTTCAAGGTGGTATTCGACACCGGATCCTCCAACCTCTGGGTGCCTTCCAAAAAGTGCCACTACACCAACATCGCTTGTTTGCTGCACAACAAGTACGACAGCCGCAAGTCCAAGACGTACGTCGCGAATGGCACCCAGTTCGCGATACAGTACGGCTCCGGCAGCCTCTCCGGCTTCCTCTCCACTGATGATGTCACCGTGGGCGGGCTCAAGGTGCGGCGCCAGACGTTCGCCGAGGCCGTGTCGGAGCCCGGGCTTGCCTTCGTGGCCGCCAAGTTCGACGGGATCCTCGGGATGGCCTTCAGCACTATCGCAGTGGACCATGTGACCCCGGTGTTCGACAACATGGTGGCTCAGGGACTCGTGCAGCCCGTCTTCTCGTTCTATCTCAACAGGGACCCCGGGGCGACAACGGGCGGCGAGTTGCTGCTGGGCGGCTCCGACCCCGCCCACTACCGAGGCGACCTCGTACGGGTGCCCCTACTCCGGGACACGTACTGGGAGTTCCACATGGACTCTGTCAATGTCAACGCTAGCCGCTTCTGTGCCCAGGGCTGCTCGGCCATCGCCGACACGGGCACGTCGCTGATCGCCGGGCCCTCCAAGGAGGTGGAAGCCCTCAACGCGGCGGTGGGCGCCACGGCCATCGCCTTCGGGCAGTACGTCGTGGACTGCAGCCTCATACCGCACCTGCCGCGAGTCACCTTTACCATCGCCGGGAACGACTTCACGCTCGAGGGCAACGACTACGTGCTCCGGGTGGCACAATTCGGGCACACGGTGTGCCTGTCCGGCTTCATGGCGCTGGACGTGCCGAAGCCCATGGGCCCGCTGTGGATCCTGGGGGACGTGTTCATCGGCAAGTACTACACGGAGTTCGACGCGGGGAACCGCCAGCTCGGGTTCGCGCCCGCCGTGTGA
Protein
MGKISLFFLALIASSVMALYRVPLHRMKTARTHFHEVGTELELLRLKYDVTGPSPEPLSNYLDAQYYGVISIGTPPQSFKVVFDTGSSNLWVPSKKCHYTNIACLLHNKYDSRKSKTYVANGTQFAIQYGSGSLSGFLSTDDVTVGGLKVRRQTFAEAVSEPGLAFVAAKFDGILGMAFSTIAVDHVTPVFDNMVAQGLVQPVFSFYLNRDPGATTGGELLLGGSDPAHYRGDLVRVPLLRDTYWEFHMDSVNVNASRFCAQGCSAIADTGTSLIAGPSKEVEALNAAVGATAIAFGQYVVDCSLIPHLPRVTFTIAGNDFTLEGNDYVLRVAQFGHTVCLSGFMALDVPKPMGPLWILGDVFIGKYYTEFDAGNRQLGFAPAV
Summary
Description
May degrade organelles involved in the biosynthesis and secretion of vitellogenin.
Subunit
Homodimer.
Similarity
Belongs to the peptidase A1 family.
Keywords
Aspartyl protease
Complete proteome
Direct protein sequencing
Disulfide bond
Glycoprotein
Hydrolase
Lysosome
Protease
Reference proteome
Signal
Zymogen
Feature
propeptide Removed in mature form
chain Lysosomal aspartic protease
chain Lysosomal aspartic protease
Uniprot
Q4U5S3
Q7Z1E4
H9JP90
A0A1V0QH26
A0A2A4JQ09
A0A1E1W6M6
+ More
A0A0N1INL3 A0A194QFM2 S4PGV2 A0A212FHV4 D6WC03 I1SSK5 A0A0T6B690 A0A182M8M9 A0A182PIB9 A0A1Q3FMT4 A0A182RHW4 A0A1V1FTJ3 V5GZE0 A0A0P4WKX9 A0A182JS20 B0XCF4 A0A084W3C8 W5JA18 A0A182JLY7 A0A182FSH0 A0A026VXU6 A0A2M4A7Z0 A0A2M4BQE4 T1DGD1 A0A2M4BQE2 A0A182N564 A0A182UVI2 A0A2M4BQE6 A0A2M3YZT8 A0A182YK81 A0A067RRR8 A0A182LCN7 A0A182IIQ3 A0A2M3YZL0 A0A2Y9D3T3 F5HIR1 A0A2M3ZE34 Q03168 W8BWQ3 A0A2J7PIK0 A0A2K7P6A4 A0A023ER88 A0A1W4WWJ7 A0A2L2Y403 A0A0K8U1U4 A0A2L2Y439 A0A0C9QXW8 A0A2I9LNV0 A0A1L8DTP4 A0A1L8DTI5 A0A182VQN5 C7AAS7 A0A182G4T3 V9N1V7 C8YZ52 A0A3S2LSS3 A0A154PHM3 A0A1L8DTI1 A0A1L8DTH6 A0A1E1WVU9 A0A140KPR6 A0A232ETM9 A0A1L8EEU1 A0A182QAK4 A0A1I8PL28 A0A0C9RFL2 A0A0A1XL62 T1PE31 A0A1B0CG28 B9X0K9 A0A2S1TY87 A0A336M4R6 F8RNZ8 A0A142G5I9 A0A0L0BPP9 A0A2P6KFM7 K9L8I5 A0A0U1X041 A0A3R7SR07 A0A0H4KC31 A0A0C9RXE2 A0A2L2Y0H5 K7J796 A0A2A3E6X5 V9IGZ8 A0A087TRE3 C3YUT2 A9U937 A0A087TX10 B3MG95 C1C0P0 A0A1A9WMP6 A0A1W4WB73 U5EX48
A0A0N1INL3 A0A194QFM2 S4PGV2 A0A212FHV4 D6WC03 I1SSK5 A0A0T6B690 A0A182M8M9 A0A182PIB9 A0A1Q3FMT4 A0A182RHW4 A0A1V1FTJ3 V5GZE0 A0A0P4WKX9 A0A182JS20 B0XCF4 A0A084W3C8 W5JA18 A0A182JLY7 A0A182FSH0 A0A026VXU6 A0A2M4A7Z0 A0A2M4BQE4 T1DGD1 A0A2M4BQE2 A0A182N564 A0A182UVI2 A0A2M4BQE6 A0A2M3YZT8 A0A182YK81 A0A067RRR8 A0A182LCN7 A0A182IIQ3 A0A2M3YZL0 A0A2Y9D3T3 F5HIR1 A0A2M3ZE34 Q03168 W8BWQ3 A0A2J7PIK0 A0A2K7P6A4 A0A023ER88 A0A1W4WWJ7 A0A2L2Y403 A0A0K8U1U4 A0A2L2Y439 A0A0C9QXW8 A0A2I9LNV0 A0A1L8DTP4 A0A1L8DTI5 A0A182VQN5 C7AAS7 A0A182G4T3 V9N1V7 C8YZ52 A0A3S2LSS3 A0A154PHM3 A0A1L8DTI1 A0A1L8DTH6 A0A1E1WVU9 A0A140KPR6 A0A232ETM9 A0A1L8EEU1 A0A182QAK4 A0A1I8PL28 A0A0C9RFL2 A0A0A1XL62 T1PE31 A0A1B0CG28 B9X0K9 A0A2S1TY87 A0A336M4R6 F8RNZ8 A0A142G5I9 A0A0L0BPP9 A0A2P6KFM7 K9L8I5 A0A0U1X041 A0A3R7SR07 A0A0H4KC31 A0A0C9RXE2 A0A2L2Y0H5 K7J796 A0A2A3E6X5 V9IGZ8 A0A087TRE3 C3YUT2 A9U937 A0A087TX10 B3MG95 C1C0P0 A0A1A9WMP6 A0A1W4WB73 U5EX48
EC Number
3.4.23.-
Pubmed
19121390
28351737
26354079
23622113
22118469
18362917
+ More
19820115 28410430 24438588 20920257 23761445 24508170 30249741 25244985 24845553 20966253 12364791 14747013 17210077 1400492 17510324 24495485 24945155 26561354 29248469 19446566 26483478 20169386 28648823 25553591 25830018 25315136 19809173 29709775 26108605 26116442 20075255 18563158 17994087
19820115 28410430 24438588 20920257 23761445 24508170 30249741 25244985 24845553 20966253 12364791 14747013 17210077 1400492 17510324 24495485 24945155 26561354 29248469 19446566 26483478 20169386 28648823 25553591 25830018 25315136 19809173 29709775 26108605 26116442 20075255 18563158 17994087
EMBL
DQ010007
AAY43135.1
AY297160
DQ417605
AAP50847.1
ABE03014.1
+ More
BABH01009676 KX827245 ARE67826.1 NWSH01000934 PCG73490.1 GDQN01008439 GDQN01001742 JAT82615.1 JAT89312.1 KQ460973 KPJ10102.1 KQ459144 KPJ03745.1 GAIX01003487 JAA89073.1 AGBW02008466 OWR53297.1 KQ971309 EEZ99098.1 JF759824 AEC03508.1 LJIG01009636 KRT82683.1 AXCM01001650 GFDL01006262 JAV28783.1 FX985412 BAX07425.1 GALX01002603 JAB65863.1 GDRN01010976 JAI67898.1 DS232688 EDS44784.1 ATLV01019917 KE525285 KFB44722.1 ADMH02001856 ETN60836.1 KK107599 QOIP01000012 EZA48592.1 RLU15934.1 GGFK01003529 MBW36850.1 GGFJ01006166 MBW55307.1 GAMD01002816 JAA98774.1 GGFJ01006164 MBW55305.1 GGFJ01006165 MBW55306.1 GGFM01001000 MBW21751.1 KK852463 KDR23365.1 APCN01002154 GGFM01000944 MBW21695.1 AAAB01008794 EAA03535.2 EGK96172.1 EGK96173.1 EGK96174.1 GGFM01006021 MBW26772.1 M95187 CH477377 GAMC01012916 JAB93639.1 NEVH01025127 PNF16139.1 GAPW01001930 JAC11668.1 IAAA01003060 LAA02891.1 GDHF01031828 JAI20486.1 IAAA01003061 LAA02898.1 GBYB01000545 JAG70312.1 GFWZ01000072 MBW20062.1 GFDF01004317 JAV09767.1 GFDF01004318 JAV09766.1 FJ360435 ACO56332.1 JXUM01042568 KQ561330 KXJ78954.1 JX508644 AGJ03549.1 FJ943775 ACV53024.1 RSAL01000241 RVE43661.1 KQ434912 KZC11376.1 GFDF01004315 JAV09769.1 GFDF01004316 JAV09768.1 GEMQ01000039 JAT91150.1 LN794623 CEO43690.1 NNAY01002254 OXU21720.1 GFDG01001552 JAV17247.1 AXCN02000174 GBXR01000060 JAG85166.1 GBXI01002188 JAD12104.1 KA646400 AFP61029.1 AJWK01010668 AB457169 BAH24176.1 MG547692 AWI66593.1 UFQT01000526 SSX24990.1 HM989013 AEI58896.1 KP262355 AMQ98967.1 JRES01001567 KNC21943.1 MWRG01013265 PRD25128.1 JF748720 AFE48185.1 KF900154 AIF27797.1 QCYY01002311 ROT71313.1 KR061518 AKO90276.1 GBYB01012571 JAG82338.1 IAAA01000183 IAAA01000184 LAA01638.1 KZ288369 PBC26791.1 JR045291 AEY59947.1 KK116397 KFM67682.1 GG666554 EEN55983.1 EF213114 ABQ10738.1 KK117137 KFM69649.1 CH902619 EDV37798.1 BT080419 ACO14843.1 GANO01001122 JAB58749.1
BABH01009676 KX827245 ARE67826.1 NWSH01000934 PCG73490.1 GDQN01008439 GDQN01001742 JAT82615.1 JAT89312.1 KQ460973 KPJ10102.1 KQ459144 KPJ03745.1 GAIX01003487 JAA89073.1 AGBW02008466 OWR53297.1 KQ971309 EEZ99098.1 JF759824 AEC03508.1 LJIG01009636 KRT82683.1 AXCM01001650 GFDL01006262 JAV28783.1 FX985412 BAX07425.1 GALX01002603 JAB65863.1 GDRN01010976 JAI67898.1 DS232688 EDS44784.1 ATLV01019917 KE525285 KFB44722.1 ADMH02001856 ETN60836.1 KK107599 QOIP01000012 EZA48592.1 RLU15934.1 GGFK01003529 MBW36850.1 GGFJ01006166 MBW55307.1 GAMD01002816 JAA98774.1 GGFJ01006164 MBW55305.1 GGFJ01006165 MBW55306.1 GGFM01001000 MBW21751.1 KK852463 KDR23365.1 APCN01002154 GGFM01000944 MBW21695.1 AAAB01008794 EAA03535.2 EGK96172.1 EGK96173.1 EGK96174.1 GGFM01006021 MBW26772.1 M95187 CH477377 GAMC01012916 JAB93639.1 NEVH01025127 PNF16139.1 GAPW01001930 JAC11668.1 IAAA01003060 LAA02891.1 GDHF01031828 JAI20486.1 IAAA01003061 LAA02898.1 GBYB01000545 JAG70312.1 GFWZ01000072 MBW20062.1 GFDF01004317 JAV09767.1 GFDF01004318 JAV09766.1 FJ360435 ACO56332.1 JXUM01042568 KQ561330 KXJ78954.1 JX508644 AGJ03549.1 FJ943775 ACV53024.1 RSAL01000241 RVE43661.1 KQ434912 KZC11376.1 GFDF01004315 JAV09769.1 GFDF01004316 JAV09768.1 GEMQ01000039 JAT91150.1 LN794623 CEO43690.1 NNAY01002254 OXU21720.1 GFDG01001552 JAV17247.1 AXCN02000174 GBXR01000060 JAG85166.1 GBXI01002188 JAD12104.1 KA646400 AFP61029.1 AJWK01010668 AB457169 BAH24176.1 MG547692 AWI66593.1 UFQT01000526 SSX24990.1 HM989013 AEI58896.1 KP262355 AMQ98967.1 JRES01001567 KNC21943.1 MWRG01013265 PRD25128.1 JF748720 AFE48185.1 KF900154 AIF27797.1 QCYY01002311 ROT71313.1 KR061518 AKO90276.1 GBYB01012571 JAG82338.1 IAAA01000183 IAAA01000184 LAA01638.1 KZ288369 PBC26791.1 JR045291 AEY59947.1 KK116397 KFM67682.1 GG666554 EEN55983.1 EF213114 ABQ10738.1 KK117137 KFM69649.1 CH902619 EDV37798.1 BT080419 ACO14843.1 GANO01001122 JAB58749.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000007266
+ More
UP000075883 UP000075885 UP000075900 UP000075881 UP000002320 UP000030765 UP000000673 UP000075880 UP000069272 UP000053097 UP000279307 UP000075884 UP000075903 UP000076408 UP000027135 UP000075882 UP000075840 UP000076407 UP000007062 UP000008820 UP000235965 UP000192223 UP000075920 UP000069940 UP000249989 UP000283053 UP000076502 UP000215335 UP000075886 UP000095300 UP000095301 UP000092461 UP000037069 UP000283509 UP000002358 UP000242457 UP000054359 UP000001554 UP000007801 UP000091820 UP000192221
UP000075883 UP000075885 UP000075900 UP000075881 UP000002320 UP000030765 UP000000673 UP000075880 UP000069272 UP000053097 UP000279307 UP000075884 UP000075903 UP000076408 UP000027135 UP000075882 UP000075840 UP000076407 UP000007062 UP000008820 UP000235965 UP000192223 UP000075920 UP000069940 UP000249989 UP000283053 UP000076502 UP000215335 UP000075886 UP000095300 UP000095301 UP000092461 UP000037069 UP000283509 UP000002358 UP000242457 UP000054359 UP000001554 UP000007801 UP000091820 UP000192221
Interpro
Gene 3D
ProteinModelPortal
Q4U5S3
Q7Z1E4
H9JP90
A0A1V0QH26
A0A2A4JQ09
A0A1E1W6M6
+ More
A0A0N1INL3 A0A194QFM2 S4PGV2 A0A212FHV4 D6WC03 I1SSK5 A0A0T6B690 A0A182M8M9 A0A182PIB9 A0A1Q3FMT4 A0A182RHW4 A0A1V1FTJ3 V5GZE0 A0A0P4WKX9 A0A182JS20 B0XCF4 A0A084W3C8 W5JA18 A0A182JLY7 A0A182FSH0 A0A026VXU6 A0A2M4A7Z0 A0A2M4BQE4 T1DGD1 A0A2M4BQE2 A0A182N564 A0A182UVI2 A0A2M4BQE6 A0A2M3YZT8 A0A182YK81 A0A067RRR8 A0A182LCN7 A0A182IIQ3 A0A2M3YZL0 A0A2Y9D3T3 F5HIR1 A0A2M3ZE34 Q03168 W8BWQ3 A0A2J7PIK0 A0A2K7P6A4 A0A023ER88 A0A1W4WWJ7 A0A2L2Y403 A0A0K8U1U4 A0A2L2Y439 A0A0C9QXW8 A0A2I9LNV0 A0A1L8DTP4 A0A1L8DTI5 A0A182VQN5 C7AAS7 A0A182G4T3 V9N1V7 C8YZ52 A0A3S2LSS3 A0A154PHM3 A0A1L8DTI1 A0A1L8DTH6 A0A1E1WVU9 A0A140KPR6 A0A232ETM9 A0A1L8EEU1 A0A182QAK4 A0A1I8PL28 A0A0C9RFL2 A0A0A1XL62 T1PE31 A0A1B0CG28 B9X0K9 A0A2S1TY87 A0A336M4R6 F8RNZ8 A0A142G5I9 A0A0L0BPP9 A0A2P6KFM7 K9L8I5 A0A0U1X041 A0A3R7SR07 A0A0H4KC31 A0A0C9RXE2 A0A2L2Y0H5 K7J796 A0A2A3E6X5 V9IGZ8 A0A087TRE3 C3YUT2 A9U937 A0A087TX10 B3MG95 C1C0P0 A0A1A9WMP6 A0A1W4WB73 U5EX48
A0A0N1INL3 A0A194QFM2 S4PGV2 A0A212FHV4 D6WC03 I1SSK5 A0A0T6B690 A0A182M8M9 A0A182PIB9 A0A1Q3FMT4 A0A182RHW4 A0A1V1FTJ3 V5GZE0 A0A0P4WKX9 A0A182JS20 B0XCF4 A0A084W3C8 W5JA18 A0A182JLY7 A0A182FSH0 A0A026VXU6 A0A2M4A7Z0 A0A2M4BQE4 T1DGD1 A0A2M4BQE2 A0A182N564 A0A182UVI2 A0A2M4BQE6 A0A2M3YZT8 A0A182YK81 A0A067RRR8 A0A182LCN7 A0A182IIQ3 A0A2M3YZL0 A0A2Y9D3T3 F5HIR1 A0A2M3ZE34 Q03168 W8BWQ3 A0A2J7PIK0 A0A2K7P6A4 A0A023ER88 A0A1W4WWJ7 A0A2L2Y403 A0A0K8U1U4 A0A2L2Y439 A0A0C9QXW8 A0A2I9LNV0 A0A1L8DTP4 A0A1L8DTI5 A0A182VQN5 C7AAS7 A0A182G4T3 V9N1V7 C8YZ52 A0A3S2LSS3 A0A154PHM3 A0A1L8DTI1 A0A1L8DTH6 A0A1E1WVU9 A0A140KPR6 A0A232ETM9 A0A1L8EEU1 A0A182QAK4 A0A1I8PL28 A0A0C9RFL2 A0A0A1XL62 T1PE31 A0A1B0CG28 B9X0K9 A0A2S1TY87 A0A336M4R6 F8RNZ8 A0A142G5I9 A0A0L0BPP9 A0A2P6KFM7 K9L8I5 A0A0U1X041 A0A3R7SR07 A0A0H4KC31 A0A0C9RXE2 A0A2L2Y0H5 K7J796 A0A2A3E6X5 V9IGZ8 A0A087TRE3 C3YUT2 A9U937 A0A087TX10 B3MG95 C1C0P0 A0A1A9WMP6 A0A1W4WB73 U5EX48
PDB
5UX4
E-value=7.26518e-110,
Score=1015
Ontologies
PATHWAY
GO
PANTHER
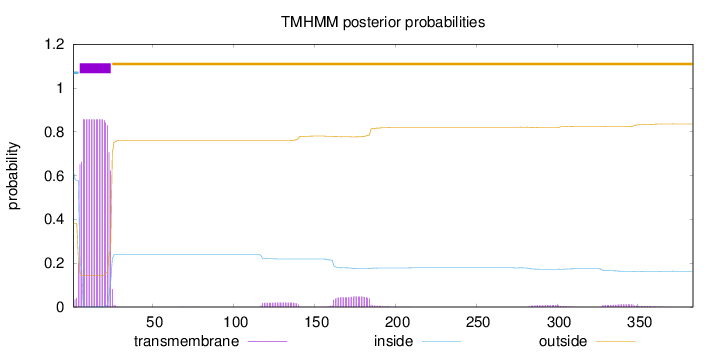
Topology
Subcellular location
Lysosome
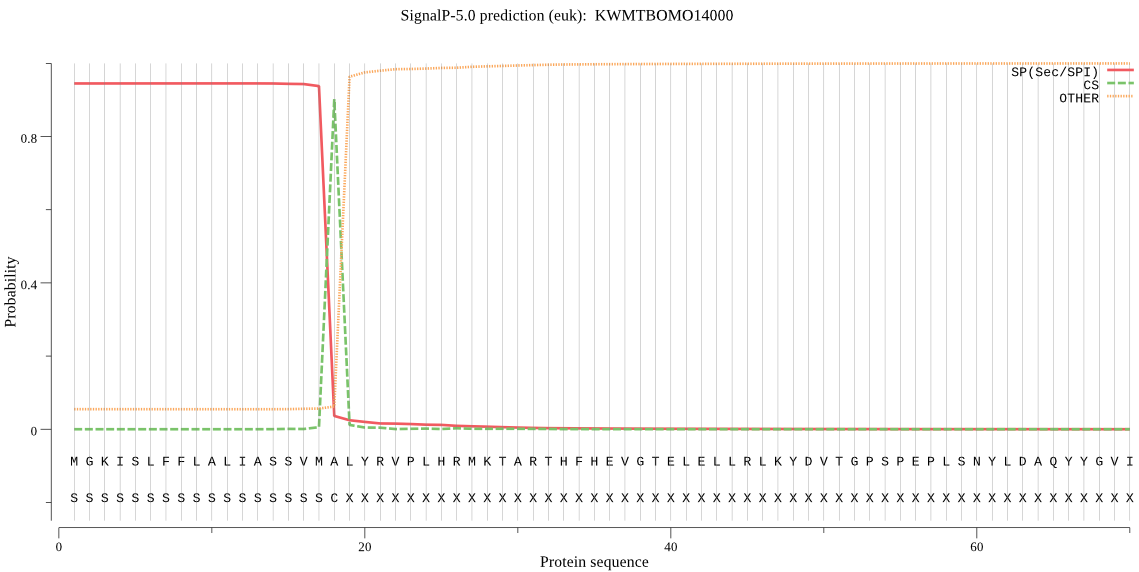
SignalP
Position: 1 - 18,
Likelihood: 0.944100
Length:
384
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.68471
Exp number, first 60 AAs:
16.67796
Total prob of N-in:
0.61737
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 24
outside
25 - 384
Population Genetic Test Statistics
Pi
204.207023
Theta
171.201316
Tajima's D
0.333796
CLR
12.138697
CSRT
0.460876956152192
Interpretation
Uncertain
