Gene
KWMTBOMO13997 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011342
Annotation
fibroinase_precursor_[Bombyx_mori]
Full name
Cathepsin L
Alternative Name
Cysteine proteinase 1
Location in the cell
Lysosomal Reliability : 2.79
Sequence
CDS
ATGAAGTGTTTAGTATTGCTGCTATGCGCAGTGGCTGCTGTGAGTGCTGTTCAGTTCTTTGACCTGGTCAAGGAAGAGTGGAGTGCCTTCAAGCTGCAGCACCGTCTCAACTACGAAAGCGAGGTCGAAGACAATTTCCGCATGAAGATATACGCTGAGCACAAGCACATCATCGCCAAACACAACCAGAAGTACGAAATGGGCCTCGTTTCTTACAAGCTGGGCATGAACAAGTACGGAGACATGCTCCACCACGAGTTCGTGAAGACTATGAACGGCTTCAACAAAACTGCCAAACACAACAAGAATCTGTACATGAAGGGTGGGAGCGTCCGCGGGGCTAAGTTCATATCGCCGGCCAACGTGAAGCTGCCGGAGCAGGTGGACTGGAGGAAGCACGGCGCCGTCACCGACATCAAGGACCAAGGGAAGTGTGGCTCATGCTGGTCCTTCAGCACGACTGGAGCTTTGGAAGGACAGCACTTCCGTCAGTCCGGCTACCTGGTGTCGCTCTCGGAGCAAAACCTCATCGACTGCTCGGAGCAGTACGGGAACAACGGCTGCAACGGGGGGCTCATGGACAACGCCTTCAAGTACATCAAGGACAACGGGGGCATCGACACCGAGCAGACCTACCCCTACGAGGGAGTTGACGACAAGTGCAGGTACAATCCCAAGAACACCGGTGCTGAGGACGTGGGCTTCGTGGACATCCCCGAGGGCGACGAACAGAAGCTGATGGAAGCCGTGGCCACCGTGGGGCCCGTCTCCGTGGCCATCGACGCCTCGCACACCAGCTTCCAGCTCTACTCCAGCGGAGTCTACAACGAGGAGGAGTGCTCCTCCACTGACCTGGACCACGGGGTTCTGGTGGTGGGTTACGGCACCGACGAGCAGGGCGTGGACTACTGGCTCGTGAAGAACTCGTGGGGCCGCTCGTGGGGCGAGCTGGGCTACATCAAGATGATCCGGAACAAGAACAACCGCTGCGGCATCGCCTCCTCCGCCTCCTACCCCCTCGTGTAA
Protein
MKCLVLLLCAVAAVSAVQFFDLVKEEWSAFKLQHRLNYESEVEDNFRMKIYAEHKHIIAKHNQKYEMGLVSYKLGMNKYGDMLHHEFVKTMNGFNKTAKHNKNLYMKGGSVRGAKFISPANVKLPEQVDWRKHGAVTDIKDQGKCGSCWSFSTTGALEGQHFRQSGYLVSLSEQNLIDCSEQYGNNGCNGGLMDNAFKYIKDNGGIDTEQTYPYEGVDDKCRYNPKNTGAEDVGFVDIPEGDEQKLMEAVATVGPVSVAIDASHTSFQLYSSGVYNEEECSSTDLDHGVLVVGYGTDEQGVDYWLVKNSWGRSWGELGYIKMIRNKNNRCGIASSASYPLV
Summary
Description
Important for the overall degradation of proteins in lysosomes. Required for differentiation of imaginal disks.
Important for the overall degradation of proteins in lysosomes. Essential for adult male and female fertility. May play a role in digestion.
Important for the overall degradation of proteins in lysosomes. Essential for adult male and female fertility. May play a role in digestion.
Catalytic Activity
Specificity close to that of papain. As compared to cathepsin B, cathepsin L exhibits higher activity toward protein substrates, but has little activity on Z-Arg-Arg-NHMec, and no peptidyl-dipeptidase activity.
Subunit
Dimer of a heavy and a light chain linked by disulfide bonds.
Similarity
Belongs to the peptidase C1 family.
Keywords
Developmental protein
Differentiation
Direct protein sequencing
Disulfide bond
Glycoprotein
Hydrolase
Lysosome
Protease
Signal
Thiol protease
Zymogen
Alternative splicing
Complete proteome
Digestion
Reference proteome
Feature
propeptide Activation peptide
chain Cathepsin L heavy chain
splice variant In isoform A.
chain Cathepsin L heavy chain
splice variant In isoform A.
Uniprot
Q6T9Z7
Q26425
C7BWY7
A0A212FHR9
A0A2A4K1J7
A0A2H1VVV9
+ More
A0A023J7N9 E2J8A8 I4DJ47 Q6UB44 F8V2Y5 A0A0N0PBH9 S4PHD4 A1DYI6 I4DM20 A0A1E1WFC2 K7J502 U5ET77 A0A194QJW0 Q26636 A0A1W4X5C5 A0A0N9ZTP4 A0A0L7RDJ1 A0A1B0EZ73 A7XH36 A0A1L8DPC9 A0A1L8DPN1 B4KTB9 A0A232F4K0 Q86FS6 C5I793 A0A0M4EVM8 D6X516 A0A1B0CQU1 Q95V59 A0A151JSU9 A0A0C9PTJ9 Q95029-2 Q95029 C6SV44 A0A1W4VQY0 A0A0L0C185 A0A088ARP1 B3NRF0 B4P445 B4JW16 A0A2A3E2D8 V9IF08 A0A0A1XMN3 A0A3B0JLP7 Q7YXL4 B4QEW3 A0A195DTS2 Q7YXL3 B4HQZ0 B4LJE9 E9IEP8 Q28XK9 B4GGA5 R4UMJ7 A0A310SMP9 A0A034W1P8 E2C9C0 O46030 A0A1V1FVL6 A0A067R0U6 V5GVR8 A0A3B0J0I7 A0A182M3E8 B3MHG5 A0A182RYY3 A0A1L8EHK8 A0A195ATU0 A0A1L8EHE3 A0A1D1ZBY8 B4MNQ5 A0A182W1X4 A0A2J7PV32 A0A0U1ZDE6 A0A182J2Y7 A0A336LNC5 A0A182QBB7 A0A158NPU2 T1PFE3 B0WI10 U4U785 N6UN71 A0A1Q3FIJ1 Q17H05 A0A182P7H9 A0A182N3W2 R4FQ93 W8BGA5 T1D4A5 A0A2M3YZP6 A0A2M4BSK6 A0A2M3YZN8 A0A1S4F2V5 J3JY43 A0A023ENZ2
A0A023J7N9 E2J8A8 I4DJ47 Q6UB44 F8V2Y5 A0A0N0PBH9 S4PHD4 A1DYI6 I4DM20 A0A1E1WFC2 K7J502 U5ET77 A0A194QJW0 Q26636 A0A1W4X5C5 A0A0N9ZTP4 A0A0L7RDJ1 A0A1B0EZ73 A7XH36 A0A1L8DPC9 A0A1L8DPN1 B4KTB9 A0A232F4K0 Q86FS6 C5I793 A0A0M4EVM8 D6X516 A0A1B0CQU1 Q95V59 A0A151JSU9 A0A0C9PTJ9 Q95029-2 Q95029 C6SV44 A0A1W4VQY0 A0A0L0C185 A0A088ARP1 B3NRF0 B4P445 B4JW16 A0A2A3E2D8 V9IF08 A0A0A1XMN3 A0A3B0JLP7 Q7YXL4 B4QEW3 A0A195DTS2 Q7YXL3 B4HQZ0 B4LJE9 E9IEP8 Q28XK9 B4GGA5 R4UMJ7 A0A310SMP9 A0A034W1P8 E2C9C0 O46030 A0A1V1FVL6 A0A067R0U6 V5GVR8 A0A3B0J0I7 A0A182M3E8 B3MHG5 A0A182RYY3 A0A1L8EHK8 A0A195ATU0 A0A1L8EHE3 A0A1D1ZBY8 B4MNQ5 A0A182W1X4 A0A2J7PV32 A0A0U1ZDE6 A0A182J2Y7 A0A336LNC5 A0A182QBB7 A0A158NPU2 T1PFE3 B0WI10 U4U785 N6UN71 A0A1Q3FIJ1 Q17H05 A0A182P7H9 A0A182N3W2 R4FQ93 W8BGA5 T1D4A5 A0A2M3YZP6 A0A2M4BSK6 A0A2M3YZN8 A0A1S4F2V5 J3JY43 A0A023ENZ2
EC Number
3.4.22.15
Pubmed
19121390
7706225
19361282
22118469
24467606
22651552
+ More
26354079 23622113 20075255 8195162 17994087 28648823 15944084 19481148 18362917 19820115 9099581 9662479 10731132 12537572 7851441 26108605 17550304 25830018 22936249 21282665 15632085 25348373 20798317 9133615 28410430 24845553 21347285 25315136 23537049 17510324 24495485 24330624 22516182 24945155
26354079 23622113 20075255 8195162 17994087 28648823 15944084 19481148 18362917 19820115 9099581 9662479 10731132 12537572 7851441 26108605 17550304 25830018 22936249 21282665 15632085 25348373 20798317 9133615 28410430 24845553 21347285 25315136 23537049 17510324 24495485 24330624 22516182 24945155
EMBL
BABH01009669
BABH01009670
AY426971
AAR87763.1
S77508
AAB33990.1
+ More
FM958000 CAX16635.1 AGBW02008466 OWR53275.1 NWSH01000235 PCG78141.1 ODYU01004593 SOQ44632.1 KC896759 AHG99253.1 HQ110065 ADN19567.1 AK401315 BAM17937.1 AY373973 AAQ75437.1 JN017204 AEJ31938.1 KQ460973 KPJ10096.1 GAIX01003317 JAA89243.1 EF068256 ABK90824.1 AK402338 BAM18960.1 GDQN01005354 GDQN01001456 JAT85700.1 JAT89598.1 GANO01002905 JAB56966.1 KQ459144 KPJ03741.1 D16533 KR534638 ALI57394.1 KQ414614 KOC68850.1 AJVK01007787 EF488012 ABR88030.1 GFDF01005778 JAV08306.1 GFDF01005779 JAV08305.1 CH933808 EDW10631.1 NNAY01000962 OXU25714.1 AY207373 AAO48766.2 FJ763762 ACR56863.1 CP012524 ALC41950.1 KQ971381 EEZ97696.1 AJWK01024035 AF426414 AAL16954.1 KQ982014 KYN30638.1 GBYB01004688 JAG74455.1 U75652 AF012089 AE013599 BT016071 D31970 BT089036 ACU12382.1 JRES01001139 KNC25214.1 CH954179 EDV56102.1 CM000158 EDW90555.1 CH916375 EDV98154.1 KZ288427 PBC25858.1 JR040710 JR040711 AEY59252.1 AEY59253.1 GBXI01002031 JAD12261.1 OUUW01000001 SPP74166.1 AY332270 KP303276 AAP94046.1 AJF94874.1 CM000362 CM002911 EDX06990.1 KMY93646.1 KQ980487 KYN15924.1 AY332271 AAP94047.1 CH480816 EDW47787.1 CH940648 EDW60529.1 GL762655 EFZ20954.1 CM000071 EAL26307.1 CH479183 EDW35525.1 KC571836 AGM32335.1 KQ760869 OAD58758.1 GAKP01010710 JAC48242.1 GL453806 EFN75465.1 D82884 BAA24442.1 FX985415 BAX07428.1 KK853019 KDR12407.1 GALX01004128 JAB64338.1 SPP74167.1 AXCM01000489 CH902619 EDV37965.2 GFDG01000710 JAV18089.1 KQ976745 KYM75472.1 GFDG01000711 JAV18088.1 GDJX01003544 JAT64392.1 CH963848 EDW73744.1 NEVH01020989 PNF20196.1 KJ913663 AJS13771.1 UFQS01002236 UFQT01000092 UFQT01002236 SSX13560.1 SSX19582.1 AXCN02001095 ADTU01022754 KA646890 AFP61519.1 DS231941 EDS28092.1 KB631815 ERL86466.1 APGK01015962 KB739708 ENN82151.1 GFDL01007699 JAV27346.1 CH477254 EAT45919.1 ACPB03002359 GAHY01000534 JAA76976.1 GAMC01006215 JAC00341.1 GALA01001028 JAA93824.1 GGFM01000986 MBW21737.1 GGFJ01006924 MBW56065.1 GGFM01000950 MBW21701.1 BT128167 AEE63128.1 GAPW01002427 GAPW01002426 JAC11172.1
FM958000 CAX16635.1 AGBW02008466 OWR53275.1 NWSH01000235 PCG78141.1 ODYU01004593 SOQ44632.1 KC896759 AHG99253.1 HQ110065 ADN19567.1 AK401315 BAM17937.1 AY373973 AAQ75437.1 JN017204 AEJ31938.1 KQ460973 KPJ10096.1 GAIX01003317 JAA89243.1 EF068256 ABK90824.1 AK402338 BAM18960.1 GDQN01005354 GDQN01001456 JAT85700.1 JAT89598.1 GANO01002905 JAB56966.1 KQ459144 KPJ03741.1 D16533 KR534638 ALI57394.1 KQ414614 KOC68850.1 AJVK01007787 EF488012 ABR88030.1 GFDF01005778 JAV08306.1 GFDF01005779 JAV08305.1 CH933808 EDW10631.1 NNAY01000962 OXU25714.1 AY207373 AAO48766.2 FJ763762 ACR56863.1 CP012524 ALC41950.1 KQ971381 EEZ97696.1 AJWK01024035 AF426414 AAL16954.1 KQ982014 KYN30638.1 GBYB01004688 JAG74455.1 U75652 AF012089 AE013599 BT016071 D31970 BT089036 ACU12382.1 JRES01001139 KNC25214.1 CH954179 EDV56102.1 CM000158 EDW90555.1 CH916375 EDV98154.1 KZ288427 PBC25858.1 JR040710 JR040711 AEY59252.1 AEY59253.1 GBXI01002031 JAD12261.1 OUUW01000001 SPP74166.1 AY332270 KP303276 AAP94046.1 AJF94874.1 CM000362 CM002911 EDX06990.1 KMY93646.1 KQ980487 KYN15924.1 AY332271 AAP94047.1 CH480816 EDW47787.1 CH940648 EDW60529.1 GL762655 EFZ20954.1 CM000071 EAL26307.1 CH479183 EDW35525.1 KC571836 AGM32335.1 KQ760869 OAD58758.1 GAKP01010710 JAC48242.1 GL453806 EFN75465.1 D82884 BAA24442.1 FX985415 BAX07428.1 KK853019 KDR12407.1 GALX01004128 JAB64338.1 SPP74167.1 AXCM01000489 CH902619 EDV37965.2 GFDG01000710 JAV18089.1 KQ976745 KYM75472.1 GFDG01000711 JAV18088.1 GDJX01003544 JAT64392.1 CH963848 EDW73744.1 NEVH01020989 PNF20196.1 KJ913663 AJS13771.1 UFQS01002236 UFQT01000092 UFQT01002236 SSX13560.1 SSX19582.1 AXCN02001095 ADTU01022754 KA646890 AFP61519.1 DS231941 EDS28092.1 KB631815 ERL86466.1 APGK01015962 KB739708 ENN82151.1 GFDL01007699 JAV27346.1 CH477254 EAT45919.1 ACPB03002359 GAHY01000534 JAA76976.1 GAMC01006215 JAC00341.1 GALA01001028 JAA93824.1 GGFM01000986 MBW21737.1 GGFJ01006924 MBW56065.1 GGFM01000950 MBW21701.1 BT128167 AEE63128.1 GAPW01002427 GAPW01002426 JAC11172.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000002358
UP000053268
+ More
UP000192223 UP000053825 UP000092462 UP000009192 UP000215335 UP000092553 UP000007266 UP000092461 UP000078541 UP000000803 UP000192221 UP000037069 UP000005203 UP000008711 UP000002282 UP000001070 UP000242457 UP000268350 UP000000304 UP000078492 UP000001292 UP000008792 UP000001819 UP000008744 UP000008237 UP000027135 UP000075883 UP000007801 UP000075900 UP000078540 UP000007798 UP000075920 UP000235965 UP000075880 UP000075886 UP000005205 UP000095301 UP000002320 UP000030742 UP000019118 UP000008820 UP000075885 UP000075884 UP000015103
UP000192223 UP000053825 UP000092462 UP000009192 UP000215335 UP000092553 UP000007266 UP000092461 UP000078541 UP000000803 UP000192221 UP000037069 UP000005203 UP000008711 UP000002282 UP000001070 UP000242457 UP000268350 UP000000304 UP000078492 UP000001292 UP000008792 UP000001819 UP000008744 UP000008237 UP000027135 UP000075883 UP000007801 UP000075900 UP000078540 UP000007798 UP000075920 UP000235965 UP000075880 UP000075886 UP000005205 UP000095301 UP000002320 UP000030742 UP000019118 UP000008820 UP000075885 UP000075884 UP000015103
Interpro
SUPFAM
SSF54001
SSF54001
ProteinModelPortal
Q6T9Z7
Q26425
C7BWY7
A0A212FHR9
A0A2A4K1J7
A0A2H1VVV9
+ More
A0A023J7N9 E2J8A8 I4DJ47 Q6UB44 F8V2Y5 A0A0N0PBH9 S4PHD4 A1DYI6 I4DM20 A0A1E1WFC2 K7J502 U5ET77 A0A194QJW0 Q26636 A0A1W4X5C5 A0A0N9ZTP4 A0A0L7RDJ1 A0A1B0EZ73 A7XH36 A0A1L8DPC9 A0A1L8DPN1 B4KTB9 A0A232F4K0 Q86FS6 C5I793 A0A0M4EVM8 D6X516 A0A1B0CQU1 Q95V59 A0A151JSU9 A0A0C9PTJ9 Q95029-2 Q95029 C6SV44 A0A1W4VQY0 A0A0L0C185 A0A088ARP1 B3NRF0 B4P445 B4JW16 A0A2A3E2D8 V9IF08 A0A0A1XMN3 A0A3B0JLP7 Q7YXL4 B4QEW3 A0A195DTS2 Q7YXL3 B4HQZ0 B4LJE9 E9IEP8 Q28XK9 B4GGA5 R4UMJ7 A0A310SMP9 A0A034W1P8 E2C9C0 O46030 A0A1V1FVL6 A0A067R0U6 V5GVR8 A0A3B0J0I7 A0A182M3E8 B3MHG5 A0A182RYY3 A0A1L8EHK8 A0A195ATU0 A0A1L8EHE3 A0A1D1ZBY8 B4MNQ5 A0A182W1X4 A0A2J7PV32 A0A0U1ZDE6 A0A182J2Y7 A0A336LNC5 A0A182QBB7 A0A158NPU2 T1PFE3 B0WI10 U4U785 N6UN71 A0A1Q3FIJ1 Q17H05 A0A182P7H9 A0A182N3W2 R4FQ93 W8BGA5 T1D4A5 A0A2M3YZP6 A0A2M4BSK6 A0A2M3YZN8 A0A1S4F2V5 J3JY43 A0A023ENZ2
A0A023J7N9 E2J8A8 I4DJ47 Q6UB44 F8V2Y5 A0A0N0PBH9 S4PHD4 A1DYI6 I4DM20 A0A1E1WFC2 K7J502 U5ET77 A0A194QJW0 Q26636 A0A1W4X5C5 A0A0N9ZTP4 A0A0L7RDJ1 A0A1B0EZ73 A7XH36 A0A1L8DPC9 A0A1L8DPN1 B4KTB9 A0A232F4K0 Q86FS6 C5I793 A0A0M4EVM8 D6X516 A0A1B0CQU1 Q95V59 A0A151JSU9 A0A0C9PTJ9 Q95029-2 Q95029 C6SV44 A0A1W4VQY0 A0A0L0C185 A0A088ARP1 B3NRF0 B4P445 B4JW16 A0A2A3E2D8 V9IF08 A0A0A1XMN3 A0A3B0JLP7 Q7YXL4 B4QEW3 A0A195DTS2 Q7YXL3 B4HQZ0 B4LJE9 E9IEP8 Q28XK9 B4GGA5 R4UMJ7 A0A310SMP9 A0A034W1P8 E2C9C0 O46030 A0A1V1FVL6 A0A067R0U6 V5GVR8 A0A3B0J0I7 A0A182M3E8 B3MHG5 A0A182RYY3 A0A1L8EHK8 A0A195ATU0 A0A1L8EHE3 A0A1D1ZBY8 B4MNQ5 A0A182W1X4 A0A2J7PV32 A0A0U1ZDE6 A0A182J2Y7 A0A336LNC5 A0A182QBB7 A0A158NPU2 T1PFE3 B0WI10 U4U785 N6UN71 A0A1Q3FIJ1 Q17H05 A0A182P7H9 A0A182N3W2 R4FQ93 W8BGA5 T1D4A5 A0A2M3YZP6 A0A2M4BSK6 A0A2M3YZN8 A0A1S4F2V5 J3JY43 A0A023ENZ2
PDB
1CS8
E-value=1.3196e-91,
Score=857
Ontologies
PATHWAY
GO
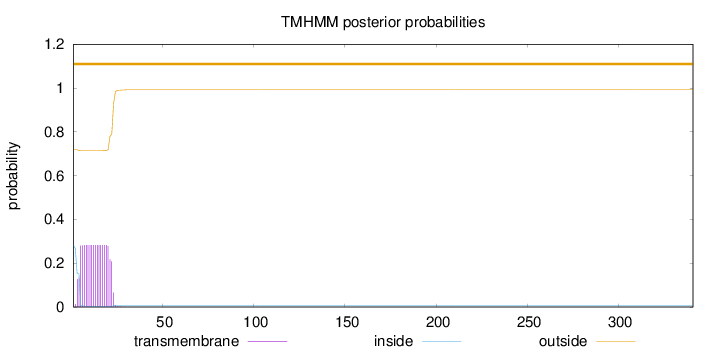
Topology
Subcellular location
Lysosome
SignalP
Position: 1 - 16,
Likelihood: 0.984480
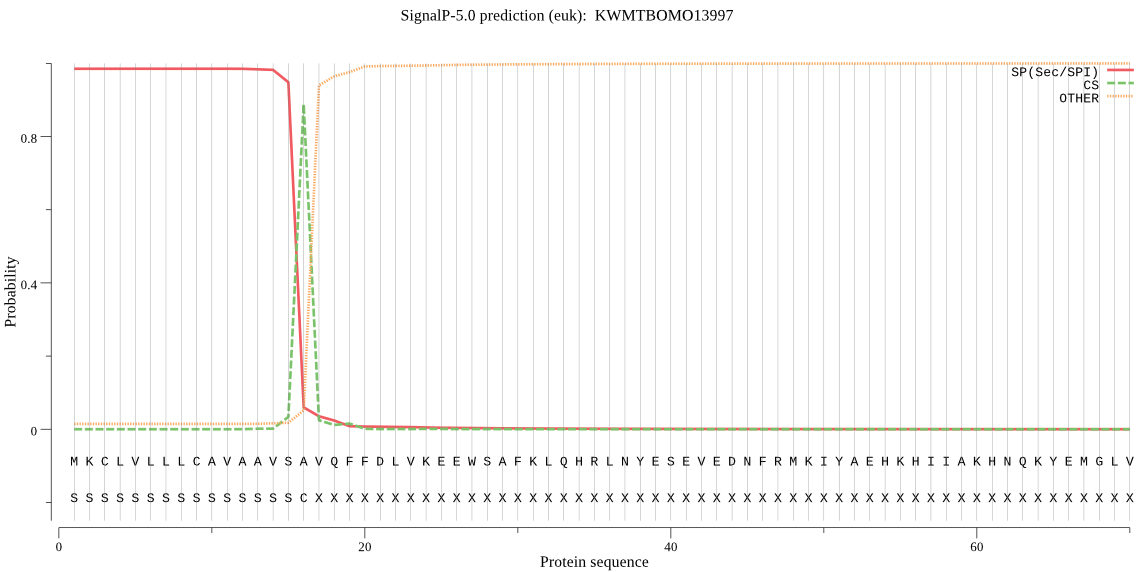
Length:
341
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.29856999999999
Exp number, first 60 AAs:
5.29828
Total prob of N-in:
0.28129
outside
1 - 341
Population Genetic Test Statistics
Pi
213.648956
Theta
180.672913
Tajima's D
0.378926
CLR
0.821101
CSRT
0.478126093695315
Interpretation
Uncertain
