Gene
KWMTBOMO13994
Pre Gene Modal
BGIBMGA011341
Annotation
PREDICTED:_syntaxin-binding_protein_5_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.782
Sequence
CDS
ATGATGATCAGCCTGCCGGAGGCCGACCTGAGGCACACGCAGCCGGTAGTCGTCTCCACAAGTGGAGGTCCCATATTTAGATTAAGAGGTTCCATACTGACGATGTCATTCCTCGACTGCAATGGTGCTTTAATTCCTTATTCATATGAAAGTTGGAAAGACGACAGTAAAGATGTTCGTGAGCGTCGGGAACGAACACCCACTAAGCAGAGTTCTTCAAGTAGTGGCAGTAGAATGAGCCCGACTCCCGGCCCCACGGAGTGCTCCTCCACAGTGGGCGACAGGCAGTTCGTAGTGATCGCCTCCGAGAAGCAGGCCAGGGTGGTGGCCTTGCCCAGTCAAAACTGTGTTTATAGACAACAAATTGTTGAAACTGATTTTATCGTTAAAGCTGAAATCGTTAGCTTGAAAGATAGTGTTTGCTTGGTCAATTACTTGTCAACCGGGCACCTGGCGGCGTACAGTCTGCCCTCACTGCGGCCTCTCATCGACGTCGACTTCCTCCCTCTCACCGAACTCAGCTTCCAGACACAGTCCAAACAGAGGGGTATCGTAGACCCAATGCTGAGTATTTGGGGCCAACAGCTCATTGTAAACGAGGACACCGATCAGATAGCGAAGACGTTCTGCTTCAGTAACCGCGGGCACGGCCTGTACCTCGCTTCGCCTTCCGAAATACAGAAGTTCACGATCGACGCTGAGTTCTGCCAACAATTGAACGAAATGTTGGGAGAAGTATTCTTGGCGCGCGACATGCCGGAGCCGCCGAAGGAGAGCTTCTTCAGGGGACTTTTCGGTGGAGGAGCGAGACCTCTTGATCGAGAGGAGCTTTTCGGGGAAAGCAGTGGAAAGGCCAGTCGCACCGTCGCCAAGCACATCCCGGGGCCAGCCTCAGGCTTGGACCAGCTCGGAGTCCGGGCCTCCACCGCTGCCAGCGAGGTCTCGCGCGCTCACCAGATGGTCGTGGAGCGCGGAGACAAGTTGTCGCAGCTCGAGGACCGCACCGAGCGGATGCACTCGCAGGCCGCCGAGTTCTCCTCCTCCGCGCACCAGCTCATGCTCAAGTACAAGGACAAGAAATGGTACCAGCTGTGA
Protein
MMISLPEADLRHTQPVVVSTSGGPIFRLRGSILTMSFLDCNGALIPYSYESWKDDSKDVRERRERTPTKQSSSSSGSRMSPTPGPTECSSTVGDRQFVVIASEKQARVVALPSQNCVYRQQIVETDFIVKAEIVSLKDSVCLVNYLSTGHLAAYSLPSLRPLIDVDFLPLTELSFQTQSKQRGIVDPMLSIWGQQLIVNEDTDQIAKTFCFSNRGHGLYLASPSEIQKFTIDAEFCQQLNEMLGEVFLARDMPEPPKESFFRGLFGGGARPLDREELFGESSGKASRTVAKHIPGPASGLDQLGVRASTAASEVSRAHQMVVERGDKLSQLEDRTERMHSQAAEFSSSAHQLMLKYKDKKWYQL
Summary
Uniprot
H9JP87
A0A212FHR0
A0A2H1VJC6
A0A2A4K293
A0A0N0PB96
A0A194QFL3
+ More
A0A1B6CZ80 W4VR65 A0A1B6CQ53 A0A3L8D4Y5 A0A139WN40 A0A1B6D2E5 V9I9N0 A0A2A3E798 A0A2J7PPT3 A0A2J7PPS3 A0A1B0CY24 A0A088A7M7 F4WAM0 E2B7M0 A0A158NPB1 A0A1Q3FYS0 A0A151WX22 A0A195AYI1 A0A195F8R5 A0A1Q3FYP2 K7J1H7 A0A2J7PPU5 E2AK15 A0A0L7R3F2 A0A2M4A1F0 Q174B3 A0A154PK40 A0A084VQC9 A0A0M8ZZR9 A0A0A1X3N2 W8AU87 F5HJT9 A0A0L7KXJ9 F5HJT8 A0A1B6LE59 A0A195DYT4 A0A182RHV3 A0A182X581 Q7PRW8 A0A182W4T4 A0A182I9Q5 A0A1A9W963 A0A1B6GVZ2 A0A182P7L8 A0A026VU24 A0A182FIJ1 A0A1B6C6I5 A0A1J1HRQ3 A0A0C9R9H7 T1PKJ5 W8B4N6 A0A232EKS1 W8BET4 A0A034VDH4 A0A1A9URJ0 T1P7V3 A0A0A1XH32 A0A034VBW4 J9JKR6 A0A1I8Q063 A0A310SHK2 A0A1I8Q049 A0A182QWE9 A0A1I8MY95 A0A2M4DP46 A0A1I8MY92 A0A1I8Q044 A0A1I8MY83 A0A2S2R9N6 A0A1B0BAY4 T1IAG2 A0A2J7PPV2 A0A2M4A134 A0A2M4A115 A0A2M3YYD1 A0A2M4CXW7 A0A182J7Y6 A0A023F534 A0A1I8Q059 A0A2J7PPU8 A0A1B6KJZ3 A0A226D8J4 A0A3Q0IVE7 A0A3B0K5H1 A0A3B0K5R3 A0A3B0K805 B4IGI1 A0A2M4CQM3 W5JLU2 A0A1I8Q047 A0A1I8MY89 Q86NY8 A0A2R7VTN1 A0A023FZ50
A0A1B6CZ80 W4VR65 A0A1B6CQ53 A0A3L8D4Y5 A0A139WN40 A0A1B6D2E5 V9I9N0 A0A2A3E798 A0A2J7PPT3 A0A2J7PPS3 A0A1B0CY24 A0A088A7M7 F4WAM0 E2B7M0 A0A158NPB1 A0A1Q3FYS0 A0A151WX22 A0A195AYI1 A0A195F8R5 A0A1Q3FYP2 K7J1H7 A0A2J7PPU5 E2AK15 A0A0L7R3F2 A0A2M4A1F0 Q174B3 A0A154PK40 A0A084VQC9 A0A0M8ZZR9 A0A0A1X3N2 W8AU87 F5HJT9 A0A0L7KXJ9 F5HJT8 A0A1B6LE59 A0A195DYT4 A0A182RHV3 A0A182X581 Q7PRW8 A0A182W4T4 A0A182I9Q5 A0A1A9W963 A0A1B6GVZ2 A0A182P7L8 A0A026VU24 A0A182FIJ1 A0A1B6C6I5 A0A1J1HRQ3 A0A0C9R9H7 T1PKJ5 W8B4N6 A0A232EKS1 W8BET4 A0A034VDH4 A0A1A9URJ0 T1P7V3 A0A0A1XH32 A0A034VBW4 J9JKR6 A0A1I8Q063 A0A310SHK2 A0A1I8Q049 A0A182QWE9 A0A1I8MY95 A0A2M4DP46 A0A1I8MY92 A0A1I8Q044 A0A1I8MY83 A0A2S2R9N6 A0A1B0BAY4 T1IAG2 A0A2J7PPV2 A0A2M4A134 A0A2M4A115 A0A2M3YYD1 A0A2M4CXW7 A0A182J7Y6 A0A023F534 A0A1I8Q059 A0A2J7PPU8 A0A1B6KJZ3 A0A226D8J4 A0A3Q0IVE7 A0A3B0K5H1 A0A3B0K5R3 A0A3B0K805 B4IGI1 A0A2M4CQM3 W5JLU2 A0A1I8Q047 A0A1I8MY89 Q86NY8 A0A2R7VTN1 A0A023FZ50
Pubmed
EMBL
BABH01009660
BABH01009661
BABH01009662
BABH01009663
AGBW02008466
OWR53280.1
+ More
ODYU01002694 SOQ40532.1 NWSH01000235 PCG78136.1 KQ460973 KPJ10095.1 KQ459144 KPJ03735.1 GEDC01018567 JAS18731.1 GANO01004111 JAB55760.1 GEDC01021778 JAS15520.1 QOIP01000014 RLU14908.1 KQ971312 KYB29277.1 GEDC01017426 JAS19872.1 JR036479 AEY57171.1 KZ288347 PBC27585.1 NEVH01022645 PNF18350.1 PNF18348.1 AJWK01035273 AJWK01035274 AJWK01035275 AJWK01035276 AJWK01035277 AJWK01035278 AJWK01035279 AJWK01035280 GL888050 EGI68778.1 GL446193 EFN88298.1 ADTU01022314 ADTU01022315 GFDL01002367 JAV32678.1 KQ982668 KYQ52489.1 KQ976703 KYM77100.1 KQ981727 KYN36771.1 GFDL01002378 JAV32667.1 PNF18352.1 GL440135 EFN66225.1 KQ414663 KOC65415.1 GGFK01001147 MBW34468.1 CH477414 EAT41412.1 KQ434943 KZC12229.1 ATLV01015189 KE525003 KFB40173.1 KQ435789 KOX74435.1 GBXI01008348 JAD05944.1 GAMC01018297 JAB88258.1 AAAB01008846 EGK96549.1 JTDY01004784 KOB67766.1 EGK96550.1 GEBQ01017999 JAT21978.1 KQ980066 KYN17887.1 EAA06458.6 APCN01001018 GECZ01003179 JAS66590.1 KK107894 EZA47308.1 GEDC01028180 JAS09118.1 CVRI01000020 CRK90735.1 GBYB01004775 JAG74542.1 KA649229 AFP63858.1 GAMC01018300 GAMC01018299 JAB88256.1 NNAY01003719 OXU18960.1 GAMC01018298 GAMC01018296 JAB88259.1 GAKP01019349 GAKP01019347 JAC39603.1 KA644622 AFP59251.1 GBXI01004509 JAD09783.1 GAKP01019350 GAKP01019348 JAC39602.1 ABLF02034698 ABLF02034701 ABLF02034702 ABLF02056744 KQ763302 OAD55129.1 AXCN02001405 GGFL01015123 MBW79301.1 GGMS01017554 MBY86757.1 JXJN01011220 JXJN01011221 JXJN01011222 ACPB03010650 ACPB03010651 ACPB03010652 PNF18349.1 GGFK01001172 MBW34493.1 GGFK01001152 MBW34473.1 GGFM01000500 MBW21251.1 GGFL01005996 MBW70174.1 GBBI01002544 JAC16168.1 PNF18351.1 GEBQ01028200 JAT11777.1 LNIX01000028 OXA41872.1 OUUW01000021 SPP89464.1 SPP89465.1 SPP89463.1 CH480836 EDW48931.1 GGFL01003401 MBW67579.1 ADMH02001203 ETN63744.1 BT003569 AAO39573.1 KK854077 PTY10816.1 GBBL01001375 JAC25945.1
ODYU01002694 SOQ40532.1 NWSH01000235 PCG78136.1 KQ460973 KPJ10095.1 KQ459144 KPJ03735.1 GEDC01018567 JAS18731.1 GANO01004111 JAB55760.1 GEDC01021778 JAS15520.1 QOIP01000014 RLU14908.1 KQ971312 KYB29277.1 GEDC01017426 JAS19872.1 JR036479 AEY57171.1 KZ288347 PBC27585.1 NEVH01022645 PNF18350.1 PNF18348.1 AJWK01035273 AJWK01035274 AJWK01035275 AJWK01035276 AJWK01035277 AJWK01035278 AJWK01035279 AJWK01035280 GL888050 EGI68778.1 GL446193 EFN88298.1 ADTU01022314 ADTU01022315 GFDL01002367 JAV32678.1 KQ982668 KYQ52489.1 KQ976703 KYM77100.1 KQ981727 KYN36771.1 GFDL01002378 JAV32667.1 PNF18352.1 GL440135 EFN66225.1 KQ414663 KOC65415.1 GGFK01001147 MBW34468.1 CH477414 EAT41412.1 KQ434943 KZC12229.1 ATLV01015189 KE525003 KFB40173.1 KQ435789 KOX74435.1 GBXI01008348 JAD05944.1 GAMC01018297 JAB88258.1 AAAB01008846 EGK96549.1 JTDY01004784 KOB67766.1 EGK96550.1 GEBQ01017999 JAT21978.1 KQ980066 KYN17887.1 EAA06458.6 APCN01001018 GECZ01003179 JAS66590.1 KK107894 EZA47308.1 GEDC01028180 JAS09118.1 CVRI01000020 CRK90735.1 GBYB01004775 JAG74542.1 KA649229 AFP63858.1 GAMC01018300 GAMC01018299 JAB88256.1 NNAY01003719 OXU18960.1 GAMC01018298 GAMC01018296 JAB88259.1 GAKP01019349 GAKP01019347 JAC39603.1 KA644622 AFP59251.1 GBXI01004509 JAD09783.1 GAKP01019350 GAKP01019348 JAC39602.1 ABLF02034698 ABLF02034701 ABLF02034702 ABLF02056744 KQ763302 OAD55129.1 AXCN02001405 GGFL01015123 MBW79301.1 GGMS01017554 MBY86757.1 JXJN01011220 JXJN01011221 JXJN01011222 ACPB03010650 ACPB03010651 ACPB03010652 PNF18349.1 GGFK01001172 MBW34493.1 GGFK01001152 MBW34473.1 GGFM01000500 MBW21251.1 GGFL01005996 MBW70174.1 GBBI01002544 JAC16168.1 PNF18351.1 GEBQ01028200 JAT11777.1 LNIX01000028 OXA41872.1 OUUW01000021 SPP89464.1 SPP89465.1 SPP89463.1 CH480836 EDW48931.1 GGFL01003401 MBW67579.1 ADMH02001203 ETN63744.1 BT003569 AAO39573.1 KK854077 PTY10816.1 GBBL01001375 JAC25945.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000053268
UP000279307
+ More
UP000007266 UP000242457 UP000235965 UP000092461 UP000005203 UP000007755 UP000008237 UP000005205 UP000075809 UP000078540 UP000078541 UP000002358 UP000000311 UP000053825 UP000008820 UP000076502 UP000030765 UP000053105 UP000007062 UP000037510 UP000078492 UP000075900 UP000076407 UP000075920 UP000075840 UP000091820 UP000075885 UP000053097 UP000069272 UP000183832 UP000215335 UP000078200 UP000007819 UP000095300 UP000075886 UP000095301 UP000092460 UP000015103 UP000075880 UP000198287 UP000079169 UP000268350 UP000001292 UP000000673
UP000007266 UP000242457 UP000235965 UP000092461 UP000005203 UP000007755 UP000008237 UP000005205 UP000075809 UP000078540 UP000078541 UP000002358 UP000000311 UP000053825 UP000008820 UP000076502 UP000030765 UP000053105 UP000007062 UP000037510 UP000078492 UP000075900 UP000076407 UP000075920 UP000075840 UP000091820 UP000075885 UP000053097 UP000069272 UP000183832 UP000215335 UP000078200 UP000007819 UP000095300 UP000075886 UP000095301 UP000092460 UP000015103 UP000075880 UP000198287 UP000079169 UP000268350 UP000001292 UP000000673
Pfam
Interpro
IPR001388
Synaptobrevin
+ More
IPR013577 LLGL2
IPR000664 Lethal2_giant
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR013905 Lgl_C_dom
IPR011048 Haem_d1_sf
IPR002939 DnaJ_C
IPR012724 DnaJ
IPR018253 DnaJ_domain_CS
IPR001305 HSP_DnaJ_Cys-rich_dom
IPR036869 J_dom_sf
IPR000863 Sulfotransferase_dom
IPR027417 P-loop_NTPase
IPR008971 HSP40/DnaJ_pept-bd
IPR001623 DnaJ_domain
IPR036410 HSP_DnaJ_Cys-rich_dom_sf
IPR017986 WD40_repeat_dom
IPR000834 Peptidase_M14
IPR008969 CarboxyPept-like_regulatory
IPR013577 LLGL2
IPR000664 Lethal2_giant
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR013905 Lgl_C_dom
IPR011048 Haem_d1_sf
IPR002939 DnaJ_C
IPR012724 DnaJ
IPR018253 DnaJ_domain_CS
IPR001305 HSP_DnaJ_Cys-rich_dom
IPR036869 J_dom_sf
IPR000863 Sulfotransferase_dom
IPR027417 P-loop_NTPase
IPR008971 HSP40/DnaJ_pept-bd
IPR001623 DnaJ_domain
IPR036410 HSP_DnaJ_Cys-rich_dom_sf
IPR017986 WD40_repeat_dom
IPR000834 Peptidase_M14
IPR008969 CarboxyPept-like_regulatory
SUPFAM
Gene 3D
ProteinModelPortal
H9JP87
A0A212FHR0
A0A2H1VJC6
A0A2A4K293
A0A0N0PB96
A0A194QFL3
+ More
A0A1B6CZ80 W4VR65 A0A1B6CQ53 A0A3L8D4Y5 A0A139WN40 A0A1B6D2E5 V9I9N0 A0A2A3E798 A0A2J7PPT3 A0A2J7PPS3 A0A1B0CY24 A0A088A7M7 F4WAM0 E2B7M0 A0A158NPB1 A0A1Q3FYS0 A0A151WX22 A0A195AYI1 A0A195F8R5 A0A1Q3FYP2 K7J1H7 A0A2J7PPU5 E2AK15 A0A0L7R3F2 A0A2M4A1F0 Q174B3 A0A154PK40 A0A084VQC9 A0A0M8ZZR9 A0A0A1X3N2 W8AU87 F5HJT9 A0A0L7KXJ9 F5HJT8 A0A1B6LE59 A0A195DYT4 A0A182RHV3 A0A182X581 Q7PRW8 A0A182W4T4 A0A182I9Q5 A0A1A9W963 A0A1B6GVZ2 A0A182P7L8 A0A026VU24 A0A182FIJ1 A0A1B6C6I5 A0A1J1HRQ3 A0A0C9R9H7 T1PKJ5 W8B4N6 A0A232EKS1 W8BET4 A0A034VDH4 A0A1A9URJ0 T1P7V3 A0A0A1XH32 A0A034VBW4 J9JKR6 A0A1I8Q063 A0A310SHK2 A0A1I8Q049 A0A182QWE9 A0A1I8MY95 A0A2M4DP46 A0A1I8MY92 A0A1I8Q044 A0A1I8MY83 A0A2S2R9N6 A0A1B0BAY4 T1IAG2 A0A2J7PPV2 A0A2M4A134 A0A2M4A115 A0A2M3YYD1 A0A2M4CXW7 A0A182J7Y6 A0A023F534 A0A1I8Q059 A0A2J7PPU8 A0A1B6KJZ3 A0A226D8J4 A0A3Q0IVE7 A0A3B0K5H1 A0A3B0K5R3 A0A3B0K805 B4IGI1 A0A2M4CQM3 W5JLU2 A0A1I8Q047 A0A1I8MY89 Q86NY8 A0A2R7VTN1 A0A023FZ50
A0A1B6CZ80 W4VR65 A0A1B6CQ53 A0A3L8D4Y5 A0A139WN40 A0A1B6D2E5 V9I9N0 A0A2A3E798 A0A2J7PPT3 A0A2J7PPS3 A0A1B0CY24 A0A088A7M7 F4WAM0 E2B7M0 A0A158NPB1 A0A1Q3FYS0 A0A151WX22 A0A195AYI1 A0A195F8R5 A0A1Q3FYP2 K7J1H7 A0A2J7PPU5 E2AK15 A0A0L7R3F2 A0A2M4A1F0 Q174B3 A0A154PK40 A0A084VQC9 A0A0M8ZZR9 A0A0A1X3N2 W8AU87 F5HJT9 A0A0L7KXJ9 F5HJT8 A0A1B6LE59 A0A195DYT4 A0A182RHV3 A0A182X581 Q7PRW8 A0A182W4T4 A0A182I9Q5 A0A1A9W963 A0A1B6GVZ2 A0A182P7L8 A0A026VU24 A0A182FIJ1 A0A1B6C6I5 A0A1J1HRQ3 A0A0C9R9H7 T1PKJ5 W8B4N6 A0A232EKS1 W8BET4 A0A034VDH4 A0A1A9URJ0 T1P7V3 A0A0A1XH32 A0A034VBW4 J9JKR6 A0A1I8Q063 A0A310SHK2 A0A1I8Q049 A0A182QWE9 A0A1I8MY95 A0A2M4DP46 A0A1I8MY92 A0A1I8Q044 A0A1I8MY83 A0A2S2R9N6 A0A1B0BAY4 T1IAG2 A0A2J7PPV2 A0A2M4A134 A0A2M4A115 A0A2M3YYD1 A0A2M4CXW7 A0A182J7Y6 A0A023F534 A0A1I8Q059 A0A2J7PPU8 A0A1B6KJZ3 A0A226D8J4 A0A3Q0IVE7 A0A3B0K5H1 A0A3B0K5R3 A0A3B0K805 B4IGI1 A0A2M4CQM3 W5JLU2 A0A1I8Q047 A0A1I8MY89 Q86NY8 A0A2R7VTN1 A0A023FZ50
PDB
1URQ
E-value=4.24134e-07,
Score=129
Ontologies
GO
GO:0016021
GO:0016192
GO:0016020
GO:0005737
GO:0005096
GO:0005886
GO:0017157
GO:0031201
GO:0019905
GO:0045159
GO:0017137
GO:0046872
GO:0031072
GO:0006457
GO:0005524
GO:0009408
GO:0008146
GO:0051082
GO:0004181
GO:0008270
GO:0006887
GO:0006893
GO:1900073
GO:0007269
GO:0007163
GO:0000149
GO:0016082
GO:0016079
GO:0008021
GO:0031594
GO:0007616
GO:0005856
GO:0016311
GO:0016791
GO:0007166
GO:0004713
GO:0016491
GO:0015930
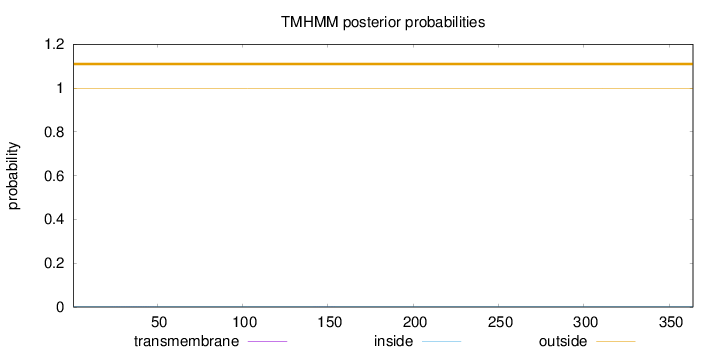
Topology
Length:
364
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0114
Exp number, first 60 AAs:
0.00895
Total prob of N-in:
0.00330
outside
1 - 364
Population Genetic Test Statistics
Pi
226.995427
Theta
210.946786
Tajima's D
0.249108
CLR
1.384373
CSRT
0.437678116094195
Interpretation
Uncertain
