Gene
KWMTBOMO13992 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011266
Annotation
sex-specific_storage-protein_1_precursor_[Bombyx_mori]
Full name
Sex-specific storage-protein 1
+ More
Basic juvenile hormone-suppressible protein 1
Basic juvenile hormone-suppressible protein 2
Basic juvenile hormone-suppressible protein 1
Basic juvenile hormone-suppressible protein 2
Alternative Name
Methionine-rich storage protein
Location in the cell
Extracellular Reliability : 3.795
Sequence
CDS
ATGAGGGTTCTAGTACTACTGGCCTGCTTGGCCGCGGCGTCAGCCTCAGCGATTAGTGGTGGCTACGGGACGATGGTCTTCACCAAGGAACCAATGGTAAACCTTGACATGAAGATGAAGGAGCTTTGCATCATGAAGCTGCTTGACCATATCCTCCAGCCGACCATGTTTGAGGACATCAAGGAGATCGCCAAGGAGTATAACATCGAGAAAAGTTGCGACAAGTACATGAATGTCGATGTCGTTAAGCAGTTCATGGAGATGTATAAGATGGGCATGCTCCCGCGTGGAGAGACCTTCGTCCACACCAACGAGCTCCAAATGGAAGAAGCCGTCAAAGTCTTCCGAGTCCTCTACTACGCTAAGGACTTCGATGTTTTCATGAGGACTGCGTGCTGGATGAGAGAAAGGATCAACGGAGGCATGTTCGTCTACGCTTTTACTGCCGCGTGCTTCCACAGAACCGACTGCAAGGGTCTCTACCTGCCCGCTCCTTACGAGATCTATCCCTACTTCTTCGTTGACAGCCATGTCATCAGTAAAGCCTTTATGATGAAGATGACTAAAGCCGCCAAGGACCCGGTCCTCTGGAAATACTACGGCATCACGGTTACTGACGACAATTTGGTAGTGATTGACTGGCGTAAGGGAGTCCGCCGCTCTTTATCCCAAAACGATGTTATGTCCTATTTCATGGAGGACGTCGACCTTAACACCTACATGTACTACCTCCACATGAACTATCCGTTCTGGATGACCGACGACGCATACGGCATAAACAAGGAGCGTCGTGGCGAGATCATGATGTACGCCAACCAGCAACTCCTAGCTAGAATGAGACTGGAACGTCTGAGCCACAAGATGTGTGACGTTAAGCCTATGATGTGGAACGAGCCCCTAGAGACCGGTTACTGGCCTAAAATCCGTCTTCCCAGCGGTGATGAAATGCCCGTCCGCCAGAACAACATGGTCGTTGCCACGAAAGATAACCTCAAGATGAAGCAGATGATGGATGATGTTGAAATGATGATCCGAGAGGGTATCCTTACTGGAAAGATCGAGCGTCGTGACGGCACTGTTATAAGCCTGAAGAAGTCTGAGGACATTGAAAACCTCGCCCGTCTGGTCCTCGGTGGCCTAGAAATTGTTGGCGACGATGCTAAAGTTATCCACTTGACCAACTTGATGAAGAAAATGCTGAGCTATGGCCAATACAACATGGACAAGTACACCTACGTGCCGACTTCCTTGGACATGTACACTACCTGTCTCCGCGATCCTGTCTTCTGGATGATCATGAAACGAGTTTGCAACATCTTCACCGTTTTCAAGAACATGCTCCCGAAATATACTCGCGAACAGTTCAGCTTCCCCGGAGTCAAAGTGGAGAAAATTACCACCGATGAGCTGGTCACATTCGTTGATGAGTACGACATGGACATTAGCAACGCTATGTACCTGGACGCAACTGAGATGCAGAACAAGACATCCGATATGACCTTTATGGCCCGCATGCGCCGCCTCAATCACCATCCGTTCCAGGTGTCGATTGATGTCATGTCCGACAAAACTGTCGATGCTGTGGTCCGCATCTTCCTGGGTCCCAAGTACGACTGCATGGGCAGGCTCATGAGCGTCAACGACAAACGCCTCGACATGTTCGAGCTGGACAGCTTTATGTACAAACTGGTCAACGGAAAGAACACCATCGTCCGTAGCTCCATGGATATGCAAGGATTTATCCCAGAGTACCTTTCGACTCGTCGTGTAATGGAAAGCGAAATGATGCCTTCCGGAGATGGCCAGACAATGGTCAAGGACTGGTGGTGCAAGTCTCGCAACGGATTCCCTCAGAGACTTATGCTGCCCCTCGGTACGATTGGAGGTTTGGAGATGCAGATGTACGTCATCGTGTCGCCGGTCCGCACCGGTATGCTCTTGCCCACATTGGACATGACCATGATGAAGGACCGTTGTGCTTGTCGCTGGAGCTCATGTATCTCCACCATGCCTCTCGGATATCCATTTGACAGACCGATTGACATGGCGAGCTTCTTCACTAACAACATGAAGTTCGCTGACGTAATGATCTACAGAAAAGACCTCGGCATGTCGAACACCAGCAAGACCGTGGACACCTCGGAGATGGTCATGATGAAGGACGATCTCACCTACCTGGACTCGGACATGCTGGTCAAGAGGACTTACAAAGACGTCATGATGATGAGCAGCATGATGAACTGA
Protein
MRVLVLLACLAAASASAISGGYGTMVFTKEPMVNLDMKMKELCIMKLLDHILQPTMFEDIKEIAKEYNIEKSCDKYMNVDVVKQFMEMYKMGMLPRGETFVHTNELQMEEAVKVFRVLYYAKDFDVFMRTACWMRERINGGMFVYAFTAACFHRTDCKGLYLPAPYEIYPYFFVDSHVISKAFMMKMTKAAKDPVLWKYYGITVTDDNLVVIDWRKGVRRSLSQNDVMSYFMEDVDLNTYMYYLHMNYPFWMTDDAYGINKERRGEIMMYANQQLLARMRLERLSHKMCDVKPMMWNEPLETGYWPKIRLPSGDEMPVRQNNMVVATKDNLKMKQMMDDVEMMIREGILTGKIERRDGTVISLKKSEDIENLARLVLGGLEIVGDDAKVIHLTNLMKKMLSYGQYNMDKYTYVPTSLDMYTTCLRDPVFWMIMKRVCNIFTVFKNMLPKYTREQFSFPGVKVEKITTDELVTFVDEYDMDISNAMYLDATEMQNKTSDMTFMARMRRLNHHPFQVSIDVMSDKTVDAVVRIFLGPKYDCMGRLMSVNDKRLDMFELDSFMYKLVNGKNTIVRSSMDMQGFIPEYLSTRRVMESEMMPSGDGQTMVKDWWCKSRNGFPQRLMLPLGTIGGLEMQMYVIVSPVRTGMLLPTLDMTMMKDRCACRWSSCISTMPLGYPFDRPIDMASFFTNNMKFADVMIYRKDLGMSNTSKTVDTSEMVMMKDDLTYLDSDMLVKRTYKDVMMMSSMMN
Summary
Similarity
Belongs to the hemocyanin family.
Keywords
Complete proteome
Glycoprotein
Reference proteome
Secreted
Signal
Storage protein
Direct protein sequencing
Feature
chain Sex-specific storage-protein 1
Uniprot
H9JP12
P09179
Q25517
Q25516
Q25518
O17481
+ More
A0A2A4JMW0 A1ILJ9 A0A3G1NI66 A0A2H1WXA7 A0A0L7LNN5 Q25050 A0A212FHR4 Q9U5K5 A0A0N1IP88 A0A194QDW4 A9LST2 A0A1U8ZSW0 Q06342 Q0WYG8 A0A0N0P9Y8 Q963S9 O77066 B7STX9 A0A1E1WT85 Q3S8M3 Q4ZJ78 D2XTA5 H9JH62 M1RNR1 A0A2W1BHW2 A0A2A4JDJ1 A2A138 A0A2H1V167 Q9U5K6 A0A194R8B2 Q963T0 A0A3G1NI75 A0A1Z2RRG6 F6MEP1 A0A212ENH9 A0A194PVL9 A2A137 A0A212ENM5 O17480 Q06343 Q9U5K7 A0A1E1WRW5 O77065 A0A2W1BMC0 Q6TG01 A0A2H1V2M0 A0A194PWA3 A0A194R7S1 A0A3G1NI74 Q9Y0E5 A0A0L7LMJ1 B0W460 A0A182H457 A0A182GR26 A0A182H055 Q16XN3 Q16I89 Q16QU8 A0A1S4FSI4 P90664 W5JSY3 A0A182FQC3 A0A182IZN6 A0A182Y3M6 T1EA57 A0A182RZ96 B0WKQ2 A0A182MT47 A0A182J7X8 W5JVA8 A0A182VLH6 A0A182VZG2 A0A1B0GLC3 Q7PXY7 A0A182WSL9 A0A182PGJ5 A0A182L9W6 A0A182UC04 A0A182HLG9 A0A182L8X1 A0A182HKM5 Q7PUR0 A0A182TGU7 A0A182X9P1 A0A1B0D919 A0A182JQ42 A0A182R4M0 A0A182UUP7 A0A182Q072 A0A182NS01 A0A182F863 T1DNH0 A0A084VS76
A0A2A4JMW0 A1ILJ9 A0A3G1NI66 A0A2H1WXA7 A0A0L7LNN5 Q25050 A0A212FHR4 Q9U5K5 A0A0N1IP88 A0A194QDW4 A9LST2 A0A1U8ZSW0 Q06342 Q0WYG8 A0A0N0P9Y8 Q963S9 O77066 B7STX9 A0A1E1WT85 Q3S8M3 Q4ZJ78 D2XTA5 H9JH62 M1RNR1 A0A2W1BHW2 A0A2A4JDJ1 A2A138 A0A2H1V167 Q9U5K6 A0A194R8B2 Q963T0 A0A3G1NI75 A0A1Z2RRG6 F6MEP1 A0A212ENH9 A0A194PVL9 A2A137 A0A212ENM5 O17480 Q06343 Q9U5K7 A0A1E1WRW5 O77065 A0A2W1BMC0 Q6TG01 A0A2H1V2M0 A0A194PWA3 A0A194R7S1 A0A3G1NI74 Q9Y0E5 A0A0L7LMJ1 B0W460 A0A182H457 A0A182GR26 A0A182H055 Q16XN3 Q16I89 Q16QU8 A0A1S4FSI4 P90664 W5JSY3 A0A182FQC3 A0A182IZN6 A0A182Y3M6 T1EA57 A0A182RZ96 B0WKQ2 A0A182MT47 A0A182J7X8 W5JVA8 A0A182VLH6 A0A182VZG2 A0A1B0GLC3 Q7PXY7 A0A182WSL9 A0A182PGJ5 A0A182L9W6 A0A182UC04 A0A182HLG9 A0A182L8X1 A0A182HKM5 Q7PUR0 A0A182TGU7 A0A182X9P1 A0A1B0D919 A0A182JQ42 A0A182R4M0 A0A182UUP7 A0A182Q072 A0A182NS01 A0A182F863 T1DNH0 A0A084VS76
Pubmed
19121390
3412900
2836399
9087538
9664700
18064702
+ More
26227816 12125057 22118469 26354079 20807423 8419330 17201777 11891129 12770171 18791259 19275945 17537657 20230828 23416133 28756777 17561427 28605547 11337253 26483478 17510324 9927174 20920257 23761445 25244985 12364791 14747013 17210077 20966253 24438588
26227816 12125057 22118469 26354079 20807423 8419330 17201777 11891129 12770171 18791259 19275945 17537657 20230828 23416133 28756777 17561427 28605547 11337253 26483478 17510324 9927174 20920257 23761445 25244985 12364791 14747013 17210077 20966253 24438588
EMBL
BABH01009655
X12978
L07610
AAA29321.1
L07609
AAA29320.1
+ More
L07611 AAA29322.1 AF032399 AAB86647.1 NWSH01000956 PCG73387.1 AB288051 BAF42698.1 KY419212 AVC68638.1 ODYU01011763 SOQ57691.1 JTDY01000468 KOB77045.1 U60988 AAB38773.1 AGBW02008466 OWR53283.1 AJ249470 CAB55604.1 KQ460541 KPJ14153.1 KQ459144 KPJ03733.1 EU259816 ABX55887.1 KU923574 ANA05288.1 L03280 AB248057 BAE98324.1 KQ459325 KPJ01439.1 AF356843 AAK71137.1 AF007768 AAC35429.1 EU711403 ACH89347.1 GDQN01000870 JAT90184.1 DQ147770 ABA02188.2 DQ004585 AAY26453.2 GU253319 ADA84299.1 BABH01012853 JX914501 AGG22606.1 KZ150082 PZC73861.1 NWSH01001869 PCG69846.1 AB266596 BAF45386.1 ODYU01000210 SOQ34603.1 AJ249469 CAB55603.1 KQ460597 KPJ13882.1 AF356842 AAK71136.1 KY419215 AVC68641.1 KY924789 ASA46443.1 JF798635 AEG19552.1 AGBW02013638 OWR43050.1 KQ459591 KPI97038.1 AB266595 BAF45385.1 OWR43051.1 AF032398 AAB86646.1 L03281 AJ249468 CAB55602.1 GDQN01001279 JAT89775.1 AF007767 AAC35428.1 PZC73860.1 AY422205 AAR32136.1 SOQ34602.1 KPI97039.1 KPJ13883.1 KY419213 AVC68639.1 AF157013 AAD39550.1 JTDY01000570 KOB76640.1 DS231835 EDS32904.1 JXUM01025392 KQ560720 KXJ81155.1 JXUM01081717 KQ563264 KXJ74179.1 JXUM01100592 KQ564582 KXJ72065.1 CH477534 EAT39409.1 CH478096 EAT33977.1 CH477731 EAT36773.1 U86080 AAB46714.1 ADMH02000246 ETN67251.1 GAMD01000727 JAB00864.1 DS231974 EDS29983.1 AXCM01007233 ETN67253.1 AJWK01034743 AAAB01008987 EAA01337.5 APCN01000900 APCN01000846 EAA01166.5 AJVK01012873 AXCN02000261 GAMD01002885 JAA98705.1 ATLV01015833 KE525036 KFB40820.1
L07611 AAA29322.1 AF032399 AAB86647.1 NWSH01000956 PCG73387.1 AB288051 BAF42698.1 KY419212 AVC68638.1 ODYU01011763 SOQ57691.1 JTDY01000468 KOB77045.1 U60988 AAB38773.1 AGBW02008466 OWR53283.1 AJ249470 CAB55604.1 KQ460541 KPJ14153.1 KQ459144 KPJ03733.1 EU259816 ABX55887.1 KU923574 ANA05288.1 L03280 AB248057 BAE98324.1 KQ459325 KPJ01439.1 AF356843 AAK71137.1 AF007768 AAC35429.1 EU711403 ACH89347.1 GDQN01000870 JAT90184.1 DQ147770 ABA02188.2 DQ004585 AAY26453.2 GU253319 ADA84299.1 BABH01012853 JX914501 AGG22606.1 KZ150082 PZC73861.1 NWSH01001869 PCG69846.1 AB266596 BAF45386.1 ODYU01000210 SOQ34603.1 AJ249469 CAB55603.1 KQ460597 KPJ13882.1 AF356842 AAK71136.1 KY419215 AVC68641.1 KY924789 ASA46443.1 JF798635 AEG19552.1 AGBW02013638 OWR43050.1 KQ459591 KPI97038.1 AB266595 BAF45385.1 OWR43051.1 AF032398 AAB86646.1 L03281 AJ249468 CAB55602.1 GDQN01001279 JAT89775.1 AF007767 AAC35428.1 PZC73860.1 AY422205 AAR32136.1 SOQ34602.1 KPI97039.1 KPJ13883.1 KY419213 AVC68639.1 AF157013 AAD39550.1 JTDY01000570 KOB76640.1 DS231835 EDS32904.1 JXUM01025392 KQ560720 KXJ81155.1 JXUM01081717 KQ563264 KXJ74179.1 JXUM01100592 KQ564582 KXJ72065.1 CH477534 EAT39409.1 CH478096 EAT33977.1 CH477731 EAT36773.1 U86080 AAB46714.1 ADMH02000246 ETN67251.1 GAMD01000727 JAB00864.1 DS231974 EDS29983.1 AXCM01007233 ETN67253.1 AJWK01034743 AAAB01008987 EAA01337.5 APCN01000900 APCN01000846 EAA01166.5 AJVK01012873 AXCN02000261 GAMD01002885 JAA98705.1 ATLV01015833 KE525036 KFB40820.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
+ More
UP000002320 UP000069940 UP000249989 UP000008820 UP000000673 UP000069272 UP000075880 UP000076408 UP000075900 UP000075883 UP000075903 UP000075920 UP000092461 UP000007062 UP000076407 UP000075885 UP000075882 UP000075902 UP000075840 UP000092462 UP000075881 UP000075886 UP000075884 UP000030765
UP000002320 UP000069940 UP000249989 UP000008820 UP000000673 UP000069272 UP000075880 UP000076408 UP000075900 UP000075883 UP000075903 UP000075920 UP000092461 UP000007062 UP000076407 UP000075885 UP000075882 UP000075902 UP000075840 UP000092462 UP000075881 UP000075886 UP000075884 UP000030765
Interpro
Gene 3D
ProteinModelPortal
H9JP12
P09179
Q25517
Q25516
Q25518
O17481
+ More
A0A2A4JMW0 A1ILJ9 A0A3G1NI66 A0A2H1WXA7 A0A0L7LNN5 Q25050 A0A212FHR4 Q9U5K5 A0A0N1IP88 A0A194QDW4 A9LST2 A0A1U8ZSW0 Q06342 Q0WYG8 A0A0N0P9Y8 Q963S9 O77066 B7STX9 A0A1E1WT85 Q3S8M3 Q4ZJ78 D2XTA5 H9JH62 M1RNR1 A0A2W1BHW2 A0A2A4JDJ1 A2A138 A0A2H1V167 Q9U5K6 A0A194R8B2 Q963T0 A0A3G1NI75 A0A1Z2RRG6 F6MEP1 A0A212ENH9 A0A194PVL9 A2A137 A0A212ENM5 O17480 Q06343 Q9U5K7 A0A1E1WRW5 O77065 A0A2W1BMC0 Q6TG01 A0A2H1V2M0 A0A194PWA3 A0A194R7S1 A0A3G1NI74 Q9Y0E5 A0A0L7LMJ1 B0W460 A0A182H457 A0A182GR26 A0A182H055 Q16XN3 Q16I89 Q16QU8 A0A1S4FSI4 P90664 W5JSY3 A0A182FQC3 A0A182IZN6 A0A182Y3M6 T1EA57 A0A182RZ96 B0WKQ2 A0A182MT47 A0A182J7X8 W5JVA8 A0A182VLH6 A0A182VZG2 A0A1B0GLC3 Q7PXY7 A0A182WSL9 A0A182PGJ5 A0A182L9W6 A0A182UC04 A0A182HLG9 A0A182L8X1 A0A182HKM5 Q7PUR0 A0A182TGU7 A0A182X9P1 A0A1B0D919 A0A182JQ42 A0A182R4M0 A0A182UUP7 A0A182Q072 A0A182NS01 A0A182F863 T1DNH0 A0A084VS76
A0A2A4JMW0 A1ILJ9 A0A3G1NI66 A0A2H1WXA7 A0A0L7LNN5 Q25050 A0A212FHR4 Q9U5K5 A0A0N1IP88 A0A194QDW4 A9LST2 A0A1U8ZSW0 Q06342 Q0WYG8 A0A0N0P9Y8 Q963S9 O77066 B7STX9 A0A1E1WT85 Q3S8M3 Q4ZJ78 D2XTA5 H9JH62 M1RNR1 A0A2W1BHW2 A0A2A4JDJ1 A2A138 A0A2H1V167 Q9U5K6 A0A194R8B2 Q963T0 A0A3G1NI75 A0A1Z2RRG6 F6MEP1 A0A212ENH9 A0A194PVL9 A2A137 A0A212ENM5 O17480 Q06343 Q9U5K7 A0A1E1WRW5 O77065 A0A2W1BMC0 Q6TG01 A0A2H1V2M0 A0A194PWA3 A0A194R7S1 A0A3G1NI74 Q9Y0E5 A0A0L7LMJ1 B0W460 A0A182H457 A0A182GR26 A0A182H055 Q16XN3 Q16I89 Q16QU8 A0A1S4FSI4 P90664 W5JSY3 A0A182FQC3 A0A182IZN6 A0A182Y3M6 T1EA57 A0A182RZ96 B0WKQ2 A0A182MT47 A0A182J7X8 W5JVA8 A0A182VLH6 A0A182VZG2 A0A1B0GLC3 Q7PXY7 A0A182WSL9 A0A182PGJ5 A0A182L9W6 A0A182UC04 A0A182HLG9 A0A182L8X1 A0A182HKM5 Q7PUR0 A0A182TGU7 A0A182X9P1 A0A1B0D919 A0A182JQ42 A0A182R4M0 A0A182UUP7 A0A182Q072 A0A182NS01 A0A182F863 T1DNH0 A0A084VS76
PDB
3WJM
E-value=4.56046e-98,
Score=916
Ontologies
PANTHER
Topology
Subcellular location
SignalP
Position: 1 - 17,
Likelihood: 0.996654
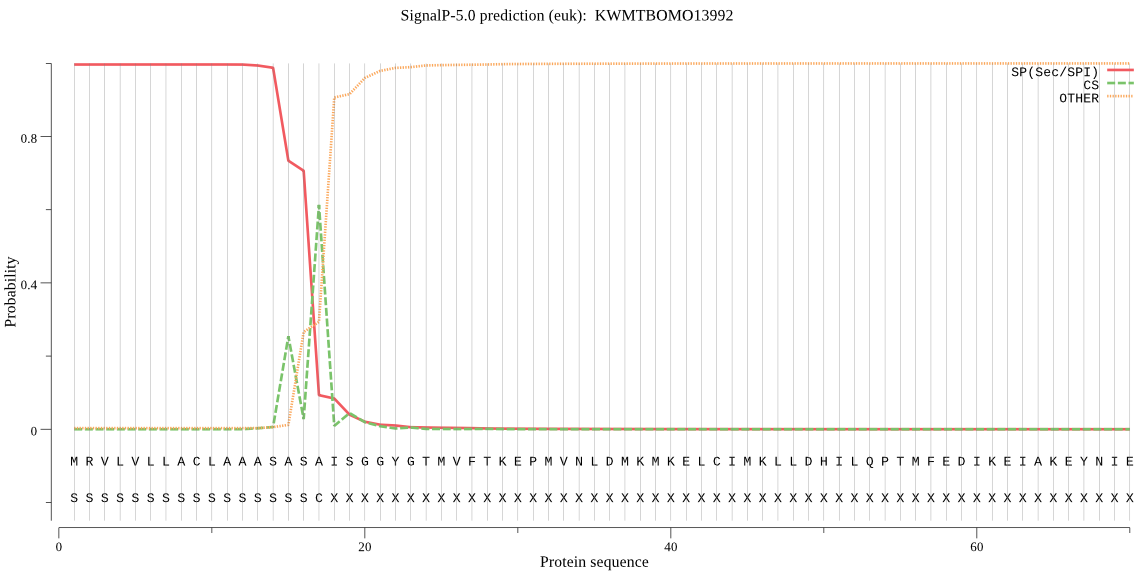
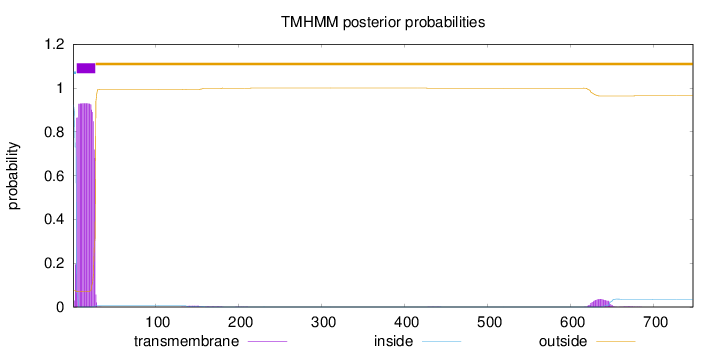
Length:
747
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.0648599999999
Exp number, first 60 AAs:
21.1274
Total prob of N-in:
0.92400
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 747
Population Genetic Test Statistics
Pi
222.379067
Theta
191.23724
Tajima's D
0.445372
CLR
1.883801
CSRT
0.496075196240188
Interpretation
Uncertain
