Gene
KWMTBOMO13991 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011268
Annotation
coproporphirynogen_oxidase_[Bombyx_mori]
Full name
Oxygen-dependent coproporphyrinogen-III oxidase
Location in the cell
Cytoplasmic Reliability : 2.565
Sequence
CDS
ATGAAAGAAATAATGCAACTTAAGAACTATATGGCAAACCCAATCACACCAGTTGAACAATTAGAGCAAAACAAAGATGATATGAAGACACAAATGGAATTACTCATCATGAGGATTCAAGCAGAATTCTGCAGGGCTTTGGAGAAAGAGGAAGACAAGGCATGGGAAGATGAAAAATCTGTGGAAGATTATGATTTTGACAATGATGTTTACATGACATGGCCGGAAGCAAAGTTTACAGTAGACAGATGGACCCGTAAAGAAGGAGGTGGTGGAATCACTTGTGTTCTACAAGACGGCAGAGTCTTTGAGAAAGCAGGAGTGAATATATCAGTTGTCTCCGGGAAACTTCCTCCAGCTGCCATTCAGCAGATGCGGAGCAGAGGCAAGAATCTACAAAACGCAGAGCTCCCGTTTTTTGCAGCCGGTGTGAGTGCCGTCATCCACCCAAGGAACCCCATGGTCCCCACCATCCACTTCAACTATAGATACTTTGAGGTCCAGGATAAGAATGGGGTCCAGTGGTGGTTCGGAGGCGGCACCGATCTGACCCCGTATTATCTGAACGAGGACGACGCTCTTCACTTCCATCTAACCTTGAAGCACGCGTGTGACGATCACGATTCATCGTATTATGCTAGATTCAAGAAATGGTGCGATGATTACTTCTACATTCCCCACCGCGGTGAAAGGCGAGGGGTCGGAGGGATATTCTTTGATGACCTGGACACTCCCAACCAGCAAGCTGCCTTCGCCTTCGTCACCTCCTGCGCTGAAGCAGTCATACCCAGCTACGTGCCTCTAGTTAAGAAGCACAAAGACGACGCTTACGGCTACCACGAGCGTCAATGGCAGTTGCTCAGACGAGGGAGATATGTGGAGTTCAATCTCATATACGACAGAGGAACAAAGTTTGGGCTGCACACTCCTGAAGCTAGATACGAATCAATACTGATGTCGCTTCCCCTTAACGCTAAATGGGAATATATGCACCAAGTCCAACCGAACTCTGAAGAAGAAAAACTGTTGAAAATCCTAAAAGAACCTAAAGATTGGCTTCAAATAGAGGATAAAAAGCGGTCTAATGCATAG
Protein
MKEIMQLKNYMANPITPVEQLEQNKDDMKTQMELLIMRIQAEFCRALEKEEDKAWEDEKSVEDYDFDNDVYMTWPEAKFTVDRWTRKEGGGGITCVLQDGRVFEKAGVNISVVSGKLPPAAIQQMRSRGKNLQNAELPFFAAGVSAVIHPRNPMVPTIHFNYRYFEVQDKNGVQWWFGGGTDLTPYYLNEDDALHFHLTLKHACDDHDSSYYARFKKWCDDYFYIPHRGERRGVGGIFFDDLDTPNQQAAFAFVTSCAEAVIPSYVPLVKKHKDDAYGYHERQWQLLRRGRYVEFNLIYDRGTKFGLHTPEARYESILMSLPLNAKWEYMHQVQPNSEEEKLLKILKEPKDWLQIEDKKRSNA
Summary
Description
Involved in the heme biosynthesis. Catalyzes the aerobic oxidative decarboxylation of propionate groups of rings A and B of coproporphyrinogen-III to yield the vinyl groups in protoporphyrinogen-IX (By similarity).
Catalytic Activity
coproporphyrinogen III + 2 H(+) + O2 = 2 CO2 + 2 H2O + protoporphyrinogen IX
Subunit
Homodimer.
Similarity
Belongs to the aerobic coproporphyrinogen-III oxidase family.
Keywords
Complete proteome
Heme biosynthesis
Oxidoreductase
Porphyrin biosynthesis
Reference proteome
Feature
chain Oxygen-dependent coproporphyrinogen-III oxidase
Uniprot
H9JP14
Q2F5S7
A0A2A4JQJ3
A0A2H1VTC1
A0A0N0PBW8
A0A2A4JRB2
+ More
A0A212FHU5 S4P7D7 A0A194QHW6 K7J6L0 A0A232F1M9 A0A151IBQ3 A0A0M8ZTB0 E9J2E8 F4WVS1 A0A195DBU7 A0A158NMQ4 A0A0L7R4T1 A0A151I0W1 A0A1B0CSC7 A0A182H450 A0A151JTZ8 A0A1B6JDU0 A0A0J7L2P3 A0A224XSD1 A0A1B6CSF4 E2ALM0 A0A0A9YSU1 Q16JA9 A0A1Y1K271 E2BSA7 A0A1L8DZJ8 A0A2J7RNP6 A0A1B6GQL4 A0A1L8E008 A0A087ZND6 A0A1B0D4Z3 A0A067RFM9 A0A2A3E615 A0A0K8V4R4 A0A034WE27 A0A2P8XXJ8 W8C3K8 A0A026WNH9 A0A023FAM8 A0A3L8DHV6 A0A0P4VQY7 T1I053 A0A1Q3F933 B0WG12 B4JPS8 A0A1I8MEW0 A0A1D2NDH9 A0A0L0BQ89 A0A1B0FEW8 D3TQ81 A0A154PFQ7 B4NZY0 A0A1W4UBM6 A0A1A9VIU2 A0A1A9Z414 U5EWJ1 B4Q5A5 B3N675 J3JTU5 N6U8J8 X2J9R2 Q9V3D2 B4LRF3 A0A182P601 A0A2S2QFD7 B4KJU1 R4WPN8 J9JNY5 A0A1I8PRG4 A0A182R2Q8 Q29JW9 A0A1A9WXG3 A0A1J1IPS8 A0A182MLR9 A0A182VF43 A0A084VZS8 A0A182QNT8 A0A182LI79 Q7Q2P4 A0A182HXH5 A0A182YI37 A0A182WZV7 A0A182TMW7 A0A182JS40 A0A182WAQ7 A0A1W4X779 A0A3B0K942 A0A182NNB7 B4MZ18 A0A182T0K3 A0A0M4EED6 B3MKT0 V4A1R5 A0A1B0BJ76
A0A212FHU5 S4P7D7 A0A194QHW6 K7J6L0 A0A232F1M9 A0A151IBQ3 A0A0M8ZTB0 E9J2E8 F4WVS1 A0A195DBU7 A0A158NMQ4 A0A0L7R4T1 A0A151I0W1 A0A1B0CSC7 A0A182H450 A0A151JTZ8 A0A1B6JDU0 A0A0J7L2P3 A0A224XSD1 A0A1B6CSF4 E2ALM0 A0A0A9YSU1 Q16JA9 A0A1Y1K271 E2BSA7 A0A1L8DZJ8 A0A2J7RNP6 A0A1B6GQL4 A0A1L8E008 A0A087ZND6 A0A1B0D4Z3 A0A067RFM9 A0A2A3E615 A0A0K8V4R4 A0A034WE27 A0A2P8XXJ8 W8C3K8 A0A026WNH9 A0A023FAM8 A0A3L8DHV6 A0A0P4VQY7 T1I053 A0A1Q3F933 B0WG12 B4JPS8 A0A1I8MEW0 A0A1D2NDH9 A0A0L0BQ89 A0A1B0FEW8 D3TQ81 A0A154PFQ7 B4NZY0 A0A1W4UBM6 A0A1A9VIU2 A0A1A9Z414 U5EWJ1 B4Q5A5 B3N675 J3JTU5 N6U8J8 X2J9R2 Q9V3D2 B4LRF3 A0A182P601 A0A2S2QFD7 B4KJU1 R4WPN8 J9JNY5 A0A1I8PRG4 A0A182R2Q8 Q29JW9 A0A1A9WXG3 A0A1J1IPS8 A0A182MLR9 A0A182VF43 A0A084VZS8 A0A182QNT8 A0A182LI79 Q7Q2P4 A0A182HXH5 A0A182YI37 A0A182WZV7 A0A182TMW7 A0A182JS40 A0A182WAQ7 A0A1W4X779 A0A3B0K942 A0A182NNB7 B4MZ18 A0A182T0K3 A0A0M4EED6 B3MKT0 V4A1R5 A0A1B0BJ76
EC Number
1.3.3.3
Pubmed
19121390
26354079
22118469
23622113
20075255
28648823
+ More
21282665 21719571 21347285 26483478 20798317 25401762 26823975 17510324 28004739 24845553 25348373 29403074 24495485 24508170 25474469 30249741 27129103 17994087 25315136 27289101 26108605 20353571 17550304 22936249 22516182 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10731138 23691247 15632085 24438588 20966253 12364791 14747013 17210077 25244985 23254933
21282665 21719571 21347285 26483478 20798317 25401762 26823975 17510324 28004739 24845553 25348373 29403074 24495485 24508170 25474469 30249741 27129103 17994087 25315136 27289101 26108605 20353571 17550304 22936249 22516182 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10731138 23691247 15632085 24438588 20966253 12364791 14747013 17210077 25244985 23254933
EMBL
BABH01009643
BABH01009644
DQ311346
ABD36290.1
NWSH01000772
PCG74307.1
+ More
ODYU01004315 SOQ44059.1 KQ460880 KPJ11526.1 PCG74306.1 AGBW02008466 OWR53287.1 GAIX01004674 JAA87886.1 KQ458761 KPJ05102.1 AAZX01001311 NNAY01001214 OXU24706.1 KQ978078 KYM97172.1 KQ435897 KOX69203.1 GL767845 EFZ13003.1 GL888394 EGI61691.1 KQ981010 KYN10312.1 ADTU01020669 KQ414657 KOC65781.1 KQ976593 KYM79650.1 AJWK01025898 JXUM01108787 KQ565264 KXJ71161.1 KQ981936 KYN32768.1 GECU01010316 JAS97390.1 LBMM01001227 KMQ96689.1 GFTR01005535 JAW10891.1 GEDC01020950 JAS16348.1 GL440609 EFN65629.1 GBHO01008325 GBRD01005130 GDHC01009278 JAG35279.1 JAG60691.1 JAQ09351.1 CH478020 EAT34350.1 GEZM01100269 GEZM01100268 JAV52927.1 GL450157 EFN81420.1 GFDF01002201 JAV11883.1 NEVH01002149 PNF42456.1 GECZ01005126 JAS64643.1 GFDF01002202 JAV11882.1 AJVK01011670 KK852498 KDR22641.1 KZ288357 PBC27150.1 GDHF01020278 GDHF01018448 GDHF01001903 GDHF01001217 JAI32036.1 JAI33866.1 JAI50411.1 JAI51097.1 GAKP01006558 GAKP01006557 GAKP01006556 JAC52395.1 PYGN01001198 PSN36732.1 GAMC01006024 JAC00532.1 KK107152 EZA57216.1 GBBI01000277 JAC18435.1 QOIP01000007 RLU20035.1 GDKW01001152 JAI55443.1 ACPB03019516 GFDL01010961 JAV24084.1 DS231922 EDS26653.1 CH916372 EDV98908.1 LJIJ01000089 ODN03026.1 JRES01001533 KNC22227.1 CCAG010000397 EZ423583 ADD19859.1 KQ434885 KZC10040.1 CM000157 EDW87807.1 GANO01002932 JAB56939.1 CM000361 CM002910 EDX04038.1 KMY88669.1 CH954177 EDV59162.1 BT126654 AEE61617.1 APGK01044452 KB741026 KB631924 ENN74927.1 ERL87148.1 AE014134 AHN54212.1 AF160897 CH940649 EDW64623.1 GGMS01007255 MBY76458.1 CH933807 EDW12544.1 AK417631 BAN20846.1 ABLF02015212 ABLF02015215 CH379062 EAL32842.1 CVRI01000057 CRL02223.1 AXCM01000075 ATLV01018955 KE525256 KFB43472.1 AXCN02001904 AAAB01008968 EAA13292.4 EDO63510.1 APCN01005305 OUUW01000006 SPP81541.1 CH963913 EDW77414.1 CP012523 ALC38480.1 CH902620 EDV31611.1 KB202953 ESO87246.1 JXJN01015341
ODYU01004315 SOQ44059.1 KQ460880 KPJ11526.1 PCG74306.1 AGBW02008466 OWR53287.1 GAIX01004674 JAA87886.1 KQ458761 KPJ05102.1 AAZX01001311 NNAY01001214 OXU24706.1 KQ978078 KYM97172.1 KQ435897 KOX69203.1 GL767845 EFZ13003.1 GL888394 EGI61691.1 KQ981010 KYN10312.1 ADTU01020669 KQ414657 KOC65781.1 KQ976593 KYM79650.1 AJWK01025898 JXUM01108787 KQ565264 KXJ71161.1 KQ981936 KYN32768.1 GECU01010316 JAS97390.1 LBMM01001227 KMQ96689.1 GFTR01005535 JAW10891.1 GEDC01020950 JAS16348.1 GL440609 EFN65629.1 GBHO01008325 GBRD01005130 GDHC01009278 JAG35279.1 JAG60691.1 JAQ09351.1 CH478020 EAT34350.1 GEZM01100269 GEZM01100268 JAV52927.1 GL450157 EFN81420.1 GFDF01002201 JAV11883.1 NEVH01002149 PNF42456.1 GECZ01005126 JAS64643.1 GFDF01002202 JAV11882.1 AJVK01011670 KK852498 KDR22641.1 KZ288357 PBC27150.1 GDHF01020278 GDHF01018448 GDHF01001903 GDHF01001217 JAI32036.1 JAI33866.1 JAI50411.1 JAI51097.1 GAKP01006558 GAKP01006557 GAKP01006556 JAC52395.1 PYGN01001198 PSN36732.1 GAMC01006024 JAC00532.1 KK107152 EZA57216.1 GBBI01000277 JAC18435.1 QOIP01000007 RLU20035.1 GDKW01001152 JAI55443.1 ACPB03019516 GFDL01010961 JAV24084.1 DS231922 EDS26653.1 CH916372 EDV98908.1 LJIJ01000089 ODN03026.1 JRES01001533 KNC22227.1 CCAG010000397 EZ423583 ADD19859.1 KQ434885 KZC10040.1 CM000157 EDW87807.1 GANO01002932 JAB56939.1 CM000361 CM002910 EDX04038.1 KMY88669.1 CH954177 EDV59162.1 BT126654 AEE61617.1 APGK01044452 KB741026 KB631924 ENN74927.1 ERL87148.1 AE014134 AHN54212.1 AF160897 CH940649 EDW64623.1 GGMS01007255 MBY76458.1 CH933807 EDW12544.1 AK417631 BAN20846.1 ABLF02015212 ABLF02015215 CH379062 EAL32842.1 CVRI01000057 CRL02223.1 AXCM01000075 ATLV01018955 KE525256 KFB43472.1 AXCN02001904 AAAB01008968 EAA13292.4 EDO63510.1 APCN01005305 OUUW01000006 SPP81541.1 CH963913 EDW77414.1 CP012523 ALC38480.1 CH902620 EDV31611.1 KB202953 ESO87246.1 JXJN01015341
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000002358
+ More
UP000215335 UP000078542 UP000053105 UP000007755 UP000078492 UP000005205 UP000053825 UP000078540 UP000092461 UP000069940 UP000249989 UP000078541 UP000036403 UP000000311 UP000008820 UP000008237 UP000235965 UP000005203 UP000092462 UP000027135 UP000242457 UP000245037 UP000053097 UP000279307 UP000015103 UP000002320 UP000001070 UP000095301 UP000094527 UP000037069 UP000092444 UP000076502 UP000002282 UP000192221 UP000078200 UP000092445 UP000000304 UP000008711 UP000019118 UP000030742 UP000000803 UP000008792 UP000075885 UP000009192 UP000007819 UP000095300 UP000075900 UP000001819 UP000091820 UP000183832 UP000075883 UP000075903 UP000030765 UP000075886 UP000075882 UP000007062 UP000075840 UP000076408 UP000076407 UP000075902 UP000075881 UP000075920 UP000192223 UP000268350 UP000075884 UP000007798 UP000075901 UP000092553 UP000007801 UP000030746 UP000092460
UP000215335 UP000078542 UP000053105 UP000007755 UP000078492 UP000005205 UP000053825 UP000078540 UP000092461 UP000069940 UP000249989 UP000078541 UP000036403 UP000000311 UP000008820 UP000008237 UP000235965 UP000005203 UP000092462 UP000027135 UP000242457 UP000245037 UP000053097 UP000279307 UP000015103 UP000002320 UP000001070 UP000095301 UP000094527 UP000037069 UP000092444 UP000076502 UP000002282 UP000192221 UP000078200 UP000092445 UP000000304 UP000008711 UP000019118 UP000030742 UP000000803 UP000008792 UP000075885 UP000009192 UP000007819 UP000095300 UP000075900 UP000001819 UP000091820 UP000183832 UP000075883 UP000075903 UP000030765 UP000075886 UP000075882 UP000007062 UP000075840 UP000076408 UP000076407 UP000075902 UP000075881 UP000075920 UP000192223 UP000268350 UP000075884 UP000007798 UP000075901 UP000092553 UP000007801 UP000030746 UP000092460
Interpro
Gene 3D
ProteinModelPortal
H9JP14
Q2F5S7
A0A2A4JQJ3
A0A2H1VTC1
A0A0N0PBW8
A0A2A4JRB2
+ More
A0A212FHU5 S4P7D7 A0A194QHW6 K7J6L0 A0A232F1M9 A0A151IBQ3 A0A0M8ZTB0 E9J2E8 F4WVS1 A0A195DBU7 A0A158NMQ4 A0A0L7R4T1 A0A151I0W1 A0A1B0CSC7 A0A182H450 A0A151JTZ8 A0A1B6JDU0 A0A0J7L2P3 A0A224XSD1 A0A1B6CSF4 E2ALM0 A0A0A9YSU1 Q16JA9 A0A1Y1K271 E2BSA7 A0A1L8DZJ8 A0A2J7RNP6 A0A1B6GQL4 A0A1L8E008 A0A087ZND6 A0A1B0D4Z3 A0A067RFM9 A0A2A3E615 A0A0K8V4R4 A0A034WE27 A0A2P8XXJ8 W8C3K8 A0A026WNH9 A0A023FAM8 A0A3L8DHV6 A0A0P4VQY7 T1I053 A0A1Q3F933 B0WG12 B4JPS8 A0A1I8MEW0 A0A1D2NDH9 A0A0L0BQ89 A0A1B0FEW8 D3TQ81 A0A154PFQ7 B4NZY0 A0A1W4UBM6 A0A1A9VIU2 A0A1A9Z414 U5EWJ1 B4Q5A5 B3N675 J3JTU5 N6U8J8 X2J9R2 Q9V3D2 B4LRF3 A0A182P601 A0A2S2QFD7 B4KJU1 R4WPN8 J9JNY5 A0A1I8PRG4 A0A182R2Q8 Q29JW9 A0A1A9WXG3 A0A1J1IPS8 A0A182MLR9 A0A182VF43 A0A084VZS8 A0A182QNT8 A0A182LI79 Q7Q2P4 A0A182HXH5 A0A182YI37 A0A182WZV7 A0A182TMW7 A0A182JS40 A0A182WAQ7 A0A1W4X779 A0A3B0K942 A0A182NNB7 B4MZ18 A0A182T0K3 A0A0M4EED6 B3MKT0 V4A1R5 A0A1B0BJ76
A0A212FHU5 S4P7D7 A0A194QHW6 K7J6L0 A0A232F1M9 A0A151IBQ3 A0A0M8ZTB0 E9J2E8 F4WVS1 A0A195DBU7 A0A158NMQ4 A0A0L7R4T1 A0A151I0W1 A0A1B0CSC7 A0A182H450 A0A151JTZ8 A0A1B6JDU0 A0A0J7L2P3 A0A224XSD1 A0A1B6CSF4 E2ALM0 A0A0A9YSU1 Q16JA9 A0A1Y1K271 E2BSA7 A0A1L8DZJ8 A0A2J7RNP6 A0A1B6GQL4 A0A1L8E008 A0A087ZND6 A0A1B0D4Z3 A0A067RFM9 A0A2A3E615 A0A0K8V4R4 A0A034WE27 A0A2P8XXJ8 W8C3K8 A0A026WNH9 A0A023FAM8 A0A3L8DHV6 A0A0P4VQY7 T1I053 A0A1Q3F933 B0WG12 B4JPS8 A0A1I8MEW0 A0A1D2NDH9 A0A0L0BQ89 A0A1B0FEW8 D3TQ81 A0A154PFQ7 B4NZY0 A0A1W4UBM6 A0A1A9VIU2 A0A1A9Z414 U5EWJ1 B4Q5A5 B3N675 J3JTU5 N6U8J8 X2J9R2 Q9V3D2 B4LRF3 A0A182P601 A0A2S2QFD7 B4KJU1 R4WPN8 J9JNY5 A0A1I8PRG4 A0A182R2Q8 Q29JW9 A0A1A9WXG3 A0A1J1IPS8 A0A182MLR9 A0A182VF43 A0A084VZS8 A0A182QNT8 A0A182LI79 Q7Q2P4 A0A182HXH5 A0A182YI37 A0A182WZV7 A0A182TMW7 A0A182JS40 A0A182WAQ7 A0A1W4X779 A0A3B0K942 A0A182NNB7 B4MZ18 A0A182T0K3 A0A0M4EED6 B3MKT0 V4A1R5 A0A1B0BJ76
PDB
2AEX
E-value=5.20773e-120,
Score=1102
Ontologies
KEGG
PATHWAY
GO
PANTHER
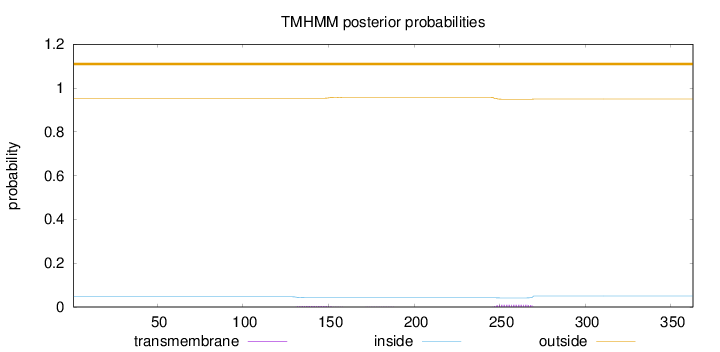
Topology
Length:
363
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29221
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04637
outside
1 - 363
Population Genetic Test Statistics
Pi
281.166643
Theta
219.959697
Tajima's D
0.889685
CLR
0.330709
CSRT
0.628818559072046
Interpretation
Uncertain
