Gene
KWMTBOMO13988 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011270
Annotation
2-oxoisovalerate_dehydrogenase_subunit_alpha?_mitochondrial_[Papilio_xuthus]
Full name
2-oxoisovalerate dehydrogenase subunit alpha
Alternative Name
Branched-chain alpha-keto acid dehydrogenase E1 component alpha chain
Location in the cell
Mitochondrial Reliability : 2.01
Sequence
CDS
ATGGCGTTGAAGACCGGAGCAAGTTTTGTCAAATTAAATGCTTTTCGACGGGCTGCTAGGCTTTTGTCGACGCGTACGAGGCCCGAAACGGCACAGAATGGCGGTGGTAAGATAGCAGAATTTCCTGGCGCAAGAGCACCGTACGTGAGTGAAATGAAATTCTTCAACGAAACTAGTTATGAGCCAATTCCGATTTATCGAGTTATGGATAATAACGGACAGATCATCGACAAAAATGAAGAACCGAACTTGGATAAGGCTACGCTGATTAACATGTACAAAACCATGGTACAACTGAGCCATATGGATAAAATTCTATATGAATCACAAAGACAAGGCCGCATTTCATTTTATATGACCAACTACGGCGAGGAGGGCATCCACATAGGCAGCGCCTCGGCACTCTCACCGAAAGATTTGGTCTTCAGTCAATACAGAGAAGTAGGAGTGTTTTTATACCGAGGGATGACTGTGACGGAACTTGTGAACCAGTGTTACGGAAACTGCGAGGATCCGGGGAAAGGGAGACAGATGCCGGTCCATTACGGAAGCAAGCACCACAACATGGTCACAATATCTAGTCCATTAGGAACACAAATGCCCCAGGCTGTTGGAGCCGCTTATGCCTACAAAAGAGTTCCGAATAACGACCGCTGCGTGATCTGCTACTTCGGTGACGGCGCTGCCTCGGAGGGCGATGCTCATGCGGCTTTCAACTTTGCCGCTACGTTAGATTGTCCTGTTATTATGCTGTGCAGAAACAACGGCTACGCGATATCTACACCCACAAGCGAACAATACCGCGGAGACGGCATCGCTTCTCGGGGACCAGCCCTGGGGCTCCACACGCTCAGAGTCGACGGGACCGACACTCTTGCGGTACACAATGCTGTGAAGAGAGCTAGAGACTTTACCGTAGCCAACAACAAGCCAGTACTCATTGAAGCTATGGCCTATCGTGTCGGGCATCATTCAACGTCTGACGATAGCAGCGCGTACAGGTCGGTGGAGGAGATACAAAAGTGGACCAAAGATGAGAGTCCTTTGCAGAAGTTCAAGCTTTATCTCGAACATAAAGGTTACTGGGACGCGGAGACTGAGAAGGCCTGGAGTAAGGAAGCAAGGGATACAGTGGTTAGAACCATGCAGGAAGCGGAAAAGAAAAAGAAACCAAACTGGAAAGAGATGCTAGAAGATGTCTACTATGAAATGCCACCCAGGCTTCAAAAACAAATGAAGCAAATGGAGGAACATCTAAAGAAGTATCCAGAGCATTACCCGCTCAATCAACACGAGAGCGATTGA
Protein
MALKTGASFVKLNAFRRAARLLSTRTRPETAQNGGGKIAEFPGARAPYVSEMKFFNETSYEPIPIYRVMDNNGQIIDKNEEPNLDKATLINMYKTMVQLSHMDKILYESQRQGRISFYMTNYGEEGIHIGSASALSPKDLVFSQYREVGVFLYRGMTVTELVNQCYGNCEDPGKGRQMPVHYGSKHHNMVTISSPLGTQMPQAVGAAYAYKRVPNNDRCVICYFGDGAASEGDAHAAFNFAATLDCPVIMLCRNNGYAISTPTSEQYRGDGIASRGPALGLHTLRVDGTDTLAVHNAVKRARDFTVANNKPVLIEAMAYRVGHHSTSDDSSAYRSVEEIQKWTKDESPLQKFKLYLEHKGYWDAETEKAWSKEARDTVVRTMQEAEKKKKPNWKEMLEDVYYEMPPRLQKQMKQMEEHLKKYPEHYPLNQHESD
Summary
Description
The branched-chain alpha-keto dehydrogenase complex catalyzes the overall conversion of alpha-keto acids to acyl-CoA and CO(2). It contains multiple copies of three enzymatic components: branched-chain alpha-keto acid decarboxylase (E1), lipoamide acyltransferase (E2) and lipoamide dehydrogenase (E3).
Catalytic Activity
3-methyl-2-oxobutanoate + [dihydrolipoyllysine-residue (2-methylpropanoyl)transferase]-(R)-N(6)-lipoyl-L-lysine + H(+) = [dihydrolipoyllysine-residue (2-methylpropanoyl)transferase]-(R)-N(6)-(S(8)-2-methylpropanoyldihydrolipoyl)-L-lysine + CO2
Cofactor
thiamine diphosphate
Similarity
Belongs to the BCKDHA family.
Feature
chain 2-oxoisovalerate dehydrogenase subunit alpha
Uniprot
H9JP16
A0A194QJL5
B2DBJ6
A0A212FHS5
C3ZXF5
W8C6P1
+ More
A0A182GJU0 E9G7A7 A0A164X3T2 W4YUZ6 A0A0P5FCW7 A0A0L0C6Y8 T1P8P0 A0A1I8PRX6 A0A1D1VN44 A0A0P4XDE8 H9G813 V4AHL2 A0A1L8F8Z5 A0A2G9SJ77 F7BVG6 B4GDS4 Q295J7 A0A0R1E2D7 B4NB33 B4PUQ4 A0A226DCN7 A0A3B0KDY4 B3NZQ5 A0A0P4WNK4 A0A1W0X2Y6 A0A0K8UZH3 A0A182PSW8 A0A1L8E0J9 Q9VHB8 A0A182QSS1 A0A1Q3F9R8 B4QWK8 C1BVS4 D6X542 A0A1W4VN32 A0A2M3Z6A5 A0A2M3Z657 T1IQP4 A0A1D2NKA3 A0A2M4BNE7 A0A182XVK2 B3M2F8 A0A182LC77 A0A0P7YCC9 A0A2M4BNB9 A0A182XE95 Q98UJ8 A0A2Y9E7P6 A0A182HJ26 A0A1W4YU45 S4R4D1 A0A182UHU6 A0A1D5NUJ3 A0A182JS64 A0A2I4CLF6 H0UTP1 A0A3Q2Y9B4 A0A2M3Z6N1 A0A1S3AKN6 B4K658 A0A182UN76 A0A3S1BQW4 A0A182LZ91 A0A2M4BNA6 A0A0B8RRX7 A0A2A2KR21 A0A210PU53 L8YCV6 A0A2A2KW39 H3BFU7 B0WYQ0 B4M4B1 A0A182NSD5 A0A1A7YB08 A0A3B4FHZ5 K1S4J4 A0A2M4BNB5 B4HJY4 I3J891 G3PHX0 A0A182W2T7 A0A3Q2WNF0 Q7PQR1 A0A0L8G146 A0A2M4AD89 A0A0M4EJ57 T1KA15 A0A3Q3FT86 A0A182FL64 A0A0D2WML6 A0A151MPF9 Q4VBU0 A0A0K2TBH9 Q16Z06
A0A182GJU0 E9G7A7 A0A164X3T2 W4YUZ6 A0A0P5FCW7 A0A0L0C6Y8 T1P8P0 A0A1I8PRX6 A0A1D1VN44 A0A0P4XDE8 H9G813 V4AHL2 A0A1L8F8Z5 A0A2G9SJ77 F7BVG6 B4GDS4 Q295J7 A0A0R1E2D7 B4NB33 B4PUQ4 A0A226DCN7 A0A3B0KDY4 B3NZQ5 A0A0P4WNK4 A0A1W0X2Y6 A0A0K8UZH3 A0A182PSW8 A0A1L8E0J9 Q9VHB8 A0A182QSS1 A0A1Q3F9R8 B4QWK8 C1BVS4 D6X542 A0A1W4VN32 A0A2M3Z6A5 A0A2M3Z657 T1IQP4 A0A1D2NKA3 A0A2M4BNE7 A0A182XVK2 B3M2F8 A0A182LC77 A0A0P7YCC9 A0A2M4BNB9 A0A182XE95 Q98UJ8 A0A2Y9E7P6 A0A182HJ26 A0A1W4YU45 S4R4D1 A0A182UHU6 A0A1D5NUJ3 A0A182JS64 A0A2I4CLF6 H0UTP1 A0A3Q2Y9B4 A0A2M3Z6N1 A0A1S3AKN6 B4K658 A0A182UN76 A0A3S1BQW4 A0A182LZ91 A0A2M4BNA6 A0A0B8RRX7 A0A2A2KR21 A0A210PU53 L8YCV6 A0A2A2KW39 H3BFU7 B0WYQ0 B4M4B1 A0A182NSD5 A0A1A7YB08 A0A3B4FHZ5 K1S4J4 A0A2M4BNB5 B4HJY4 I3J891 G3PHX0 A0A182W2T7 A0A3Q2WNF0 Q7PQR1 A0A0L8G146 A0A2M4AD89 A0A0M4EJ57 T1KA15 A0A3Q3FT86 A0A182FL64 A0A0D2WML6 A0A151MPF9 Q4VBU0 A0A0K2TBH9 Q16Z06
EC Number
1.2.4.4
Pubmed
19121390
26354079
18712529
22118469
18563158
24495485
+ More
26483478 21292972 26108605 25315136 27649274 21881562 23254933 27762356 20431018 17994087 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 27289101 25244985 20966253 11168412 15592404 21993624 25476704 28812685 23385571 9215903 22992520 25186727 12364791 14747013 17210077 22293439 23594743 17510324
26483478 21292972 26108605 25315136 27649274 21881562 23254933 27762356 20431018 17994087 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 27289101 25244985 20966253 11168412 15592404 21993624 25476704 28812685 23385571 9215903 22992520 25186727 12364791 14747013 17210077 22293439 23594743 17510324
EMBL
BABH01009639
KQ458761
KPJ05105.1
AB264679
BAG30755.1
AGBW02008466
+ More
OWR53290.1 GG666711 EEN42780.1 GAMC01004131 GAMC01004129 JAC02427.1 JXUM01068836 KQ562520 KXJ75720.1 GL732534 EFX84437.1 LRGB01001019 KZS13846.1 AAGJ04083947 AAGJ04083948 AAGJ04083949 AAGJ04083950 AAGJ04083951 AAGJ04083952 AAGJ04083953 AAGJ04083954 GDIQ01257318 JAJ94406.1 JRES01000819 KNC28173.1 KA645044 AFP59673.1 BDGG01000009 GAV03032.1 GDIP01245806 JAI77595.1 KB201755 ESO94690.1 CM004480 OCT68053.1 KV924665 PIO40165.1 AAMC01092292 CH479182 EDW34585.1 CM000070 EAL28714.1 CM000160 KRK03289.1 CH964232 EDW80997.1 EDW96671.1 LNIX01000025 OXA42594.1 OUUW01000008 SPP83886.1 CH954181 EDV49765.1 GDRN01057381 JAI65697.1 MTYJ01000021 OQV21811.1 GDHF01020250 JAI32064.1 GFDF01001867 JAV12217.1 AE014297 AY051542 AAF54398.1 AAK92966.1 AXCN02000238 GFDL01010786 JAV24259.1 CM000364 EDX13612.1 BT078703 ACO13127.1 KQ971382 EEZ97198.1 GGFM01003302 MBW24053.1 GGFM01003266 MBW24017.1 JH431312 LJIJ01000018 ODN05713.1 GGFJ01005434 MBW54575.1 CH902617 EDV43411.1 JARO02007511 KPP63866.1 GGFJ01005429 MBW54570.1 AB035083 BAB32665.1 APCN01002118 AADN05000777 AAKN02054853 GGFM01003403 MBW24154.1 CH933806 EDW14108.1 RQTK01000135 RUS86565.1 AXCM01000247 GGFJ01005428 MBW54569.1 GBSH01000125 JAG68899.1 LIAE01007889 PAV76395.1 NEDP02005491 OWF40037.1 KB364659 ELV12201.1 LIAE01007609 PAV78138.1 AFYH01010055 DS232196 EDS37160.1 CH940652 EDW59472.1 HADW01003901 HADX01005087 SBP27319.1 JH816160 EKC42336.1 GGFJ01005435 MBW54576.1 CH480815 EDW42868.1 AERX01055380 AAAB01008879 EAA08445.6 KQ424627 KOF70732.1 GGFK01005418 MBW38739.1 CP012526 ALC46871.1 CAEY01001893 KE346362 KJE91448.1 AKHW03005571 KYO26309.1 BX247864 BX649594 BC095157 BC164675 AAH95157.1 AAI64675.1 HACA01005464 HACA01005465 CDW22826.1 CH477505 EAT39862.1
OWR53290.1 GG666711 EEN42780.1 GAMC01004131 GAMC01004129 JAC02427.1 JXUM01068836 KQ562520 KXJ75720.1 GL732534 EFX84437.1 LRGB01001019 KZS13846.1 AAGJ04083947 AAGJ04083948 AAGJ04083949 AAGJ04083950 AAGJ04083951 AAGJ04083952 AAGJ04083953 AAGJ04083954 GDIQ01257318 JAJ94406.1 JRES01000819 KNC28173.1 KA645044 AFP59673.1 BDGG01000009 GAV03032.1 GDIP01245806 JAI77595.1 KB201755 ESO94690.1 CM004480 OCT68053.1 KV924665 PIO40165.1 AAMC01092292 CH479182 EDW34585.1 CM000070 EAL28714.1 CM000160 KRK03289.1 CH964232 EDW80997.1 EDW96671.1 LNIX01000025 OXA42594.1 OUUW01000008 SPP83886.1 CH954181 EDV49765.1 GDRN01057381 JAI65697.1 MTYJ01000021 OQV21811.1 GDHF01020250 JAI32064.1 GFDF01001867 JAV12217.1 AE014297 AY051542 AAF54398.1 AAK92966.1 AXCN02000238 GFDL01010786 JAV24259.1 CM000364 EDX13612.1 BT078703 ACO13127.1 KQ971382 EEZ97198.1 GGFM01003302 MBW24053.1 GGFM01003266 MBW24017.1 JH431312 LJIJ01000018 ODN05713.1 GGFJ01005434 MBW54575.1 CH902617 EDV43411.1 JARO02007511 KPP63866.1 GGFJ01005429 MBW54570.1 AB035083 BAB32665.1 APCN01002118 AADN05000777 AAKN02054853 GGFM01003403 MBW24154.1 CH933806 EDW14108.1 RQTK01000135 RUS86565.1 AXCM01000247 GGFJ01005428 MBW54569.1 GBSH01000125 JAG68899.1 LIAE01007889 PAV76395.1 NEDP02005491 OWF40037.1 KB364659 ELV12201.1 LIAE01007609 PAV78138.1 AFYH01010055 DS232196 EDS37160.1 CH940652 EDW59472.1 HADW01003901 HADX01005087 SBP27319.1 JH816160 EKC42336.1 GGFJ01005435 MBW54576.1 CH480815 EDW42868.1 AERX01055380 AAAB01008879 EAA08445.6 KQ424627 KOF70732.1 GGFK01005418 MBW38739.1 CP012526 ALC46871.1 CAEY01001893 KE346362 KJE91448.1 AKHW03005571 KYO26309.1 BX247864 BX649594 BC095157 BC164675 AAH95157.1 AAI64675.1 HACA01005464 HACA01005465 CDW22826.1 CH477505 EAT39862.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000001554
UP000069940
UP000249989
+ More
UP000000305 UP000076858 UP000007110 UP000037069 UP000095301 UP000095300 UP000186922 UP000001646 UP000030746 UP000186698 UP000008143 UP000008744 UP000001819 UP000002282 UP000007798 UP000198287 UP000268350 UP000008711 UP000075885 UP000000803 UP000075886 UP000000304 UP000007266 UP000192221 UP000094527 UP000076408 UP000007801 UP000075882 UP000034805 UP000076407 UP000248480 UP000075840 UP000192224 UP000245300 UP000075902 UP000000539 UP000075881 UP000192220 UP000005447 UP000264820 UP000079721 UP000009192 UP000075903 UP000271974 UP000075883 UP000218231 UP000242188 UP000011518 UP000008672 UP000002320 UP000008792 UP000075884 UP000261460 UP000005408 UP000001292 UP000005207 UP000007635 UP000075920 UP000264840 UP000007062 UP000053454 UP000092553 UP000015104 UP000261660 UP000069272 UP000008743 UP000050525 UP000000437 UP000008820
UP000000305 UP000076858 UP000007110 UP000037069 UP000095301 UP000095300 UP000186922 UP000001646 UP000030746 UP000186698 UP000008143 UP000008744 UP000001819 UP000002282 UP000007798 UP000198287 UP000268350 UP000008711 UP000075885 UP000000803 UP000075886 UP000000304 UP000007266 UP000192221 UP000094527 UP000076408 UP000007801 UP000075882 UP000034805 UP000076407 UP000248480 UP000075840 UP000192224 UP000245300 UP000075902 UP000000539 UP000075881 UP000192220 UP000005447 UP000264820 UP000079721 UP000009192 UP000075903 UP000271974 UP000075883 UP000218231 UP000242188 UP000011518 UP000008672 UP000002320 UP000008792 UP000075884 UP000261460 UP000005408 UP000001292 UP000005207 UP000007635 UP000075920 UP000264840 UP000007062 UP000053454 UP000092553 UP000015104 UP000261660 UP000069272 UP000008743 UP000050525 UP000000437 UP000008820
Interpro
Gene 3D
ProteinModelPortal
H9JP16
A0A194QJL5
B2DBJ6
A0A212FHS5
C3ZXF5
W8C6P1
+ More
A0A182GJU0 E9G7A7 A0A164X3T2 W4YUZ6 A0A0P5FCW7 A0A0L0C6Y8 T1P8P0 A0A1I8PRX6 A0A1D1VN44 A0A0P4XDE8 H9G813 V4AHL2 A0A1L8F8Z5 A0A2G9SJ77 F7BVG6 B4GDS4 Q295J7 A0A0R1E2D7 B4NB33 B4PUQ4 A0A226DCN7 A0A3B0KDY4 B3NZQ5 A0A0P4WNK4 A0A1W0X2Y6 A0A0K8UZH3 A0A182PSW8 A0A1L8E0J9 Q9VHB8 A0A182QSS1 A0A1Q3F9R8 B4QWK8 C1BVS4 D6X542 A0A1W4VN32 A0A2M3Z6A5 A0A2M3Z657 T1IQP4 A0A1D2NKA3 A0A2M4BNE7 A0A182XVK2 B3M2F8 A0A182LC77 A0A0P7YCC9 A0A2M4BNB9 A0A182XE95 Q98UJ8 A0A2Y9E7P6 A0A182HJ26 A0A1W4YU45 S4R4D1 A0A182UHU6 A0A1D5NUJ3 A0A182JS64 A0A2I4CLF6 H0UTP1 A0A3Q2Y9B4 A0A2M3Z6N1 A0A1S3AKN6 B4K658 A0A182UN76 A0A3S1BQW4 A0A182LZ91 A0A2M4BNA6 A0A0B8RRX7 A0A2A2KR21 A0A210PU53 L8YCV6 A0A2A2KW39 H3BFU7 B0WYQ0 B4M4B1 A0A182NSD5 A0A1A7YB08 A0A3B4FHZ5 K1S4J4 A0A2M4BNB5 B4HJY4 I3J891 G3PHX0 A0A182W2T7 A0A3Q2WNF0 Q7PQR1 A0A0L8G146 A0A2M4AD89 A0A0M4EJ57 T1KA15 A0A3Q3FT86 A0A182FL64 A0A0D2WML6 A0A151MPF9 Q4VBU0 A0A0K2TBH9 Q16Z06
A0A182GJU0 E9G7A7 A0A164X3T2 W4YUZ6 A0A0P5FCW7 A0A0L0C6Y8 T1P8P0 A0A1I8PRX6 A0A1D1VN44 A0A0P4XDE8 H9G813 V4AHL2 A0A1L8F8Z5 A0A2G9SJ77 F7BVG6 B4GDS4 Q295J7 A0A0R1E2D7 B4NB33 B4PUQ4 A0A226DCN7 A0A3B0KDY4 B3NZQ5 A0A0P4WNK4 A0A1W0X2Y6 A0A0K8UZH3 A0A182PSW8 A0A1L8E0J9 Q9VHB8 A0A182QSS1 A0A1Q3F9R8 B4QWK8 C1BVS4 D6X542 A0A1W4VN32 A0A2M3Z6A5 A0A2M3Z657 T1IQP4 A0A1D2NKA3 A0A2M4BNE7 A0A182XVK2 B3M2F8 A0A182LC77 A0A0P7YCC9 A0A2M4BNB9 A0A182XE95 Q98UJ8 A0A2Y9E7P6 A0A182HJ26 A0A1W4YU45 S4R4D1 A0A182UHU6 A0A1D5NUJ3 A0A182JS64 A0A2I4CLF6 H0UTP1 A0A3Q2Y9B4 A0A2M3Z6N1 A0A1S3AKN6 B4K658 A0A182UN76 A0A3S1BQW4 A0A182LZ91 A0A2M4BNA6 A0A0B8RRX7 A0A2A2KR21 A0A210PU53 L8YCV6 A0A2A2KW39 H3BFU7 B0WYQ0 B4M4B1 A0A182NSD5 A0A1A7YB08 A0A3B4FHZ5 K1S4J4 A0A2M4BNB5 B4HJY4 I3J891 G3PHX0 A0A182W2T7 A0A3Q2WNF0 Q7PQR1 A0A0L8G146 A0A2M4AD89 A0A0M4EJ57 T1KA15 A0A3Q3FT86 A0A182FL64 A0A0D2WML6 A0A151MPF9 Q4VBU0 A0A0K2TBH9 Q16Z06
PDB
2BEW
E-value=2.23835e-138,
Score=1262
Ontologies
PATHWAY
GO
PANTHER
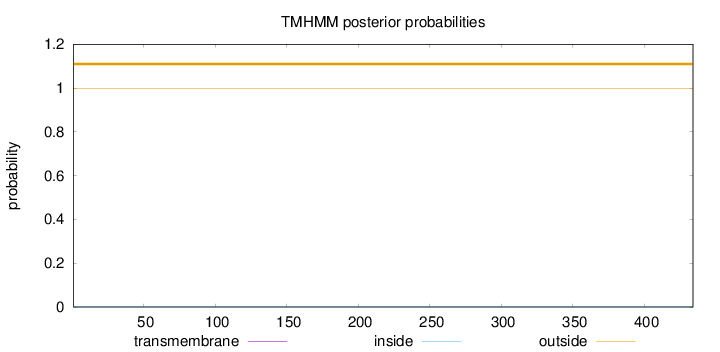
Topology
Length:
434
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00167
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00257
outside
1 - 434
Population Genetic Test Statistics
Pi
217.513308
Theta
147.846892
Tajima's D
0.072049
CLR
0.718342
CSRT
0.388330583470826
Interpretation
Uncertain
