Gene
KWMTBOMO13987
Pre Gene Modal
BGIBMGA011270
Annotation
PREDICTED:_WD_repeat-containing_protein_6_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.962
Sequence
CDS
ATGTCTTTATTAATACGTACAGATGTTACGGCTGTTAAAATTTTCAAAAACTACGTTCTTGCCGGTATTGGAAGCACTATAAATGTATTCGATAAAGCTTCGGCGAGATTAATTACGAAGTTTCAATGTTTAAAAAGCCAAAAAATATATGGTTTTGTAGTATCTAATTGCGGTAATAAAATACTCGTGTTTGGCGGAAAACAGTTCACAGTTTTATCTTTTACGCACTCGCATTATGAGGAGACATTTAAGAGAAACTTCGAACCTATAATATGCGATGATTGGCTTCATTCCGGAGTTTGGCTGAAAGAAGATATTGTTATTTTATTAACTGCTCATAATGTTACTCAGAAATGGAACTTATCAACACAGTCCTTCATATCTCAAGAAGCAGGCAAAGAAAACTCAATATTATACAGTGGACTTATTGTAATGCTGCAAGATGCAATTATGGTCCTAGCTGGGACTGTGTTCTCAGATGTCATCGTGCATAGTAATACTGTTAAGGAACCTCTACATTATTTGAGAGGACACAAGGGTGTCATATTCTCCATATCGTGTGATACAGATAAAGGTTTAATAGTAACAACTTCAGATGACAGATCAGTCAGGATTTGGAGCACAAATATCATAACAGCAAATGCGCATATGCTACAATATTGGAAGGACCATGAAATAACATGCAAATATGAACTGTACGGGCATTCGGCACGAGTCATGAGAAACTGCATCACAAATAATATAGTTATATCAGTAGGAGAAGATTCCGCTATATGCTTTTGGAATTTGAATGGAGAACTAATAAAAAAGAATAACTCACATCAGTATTCTGGTATATGGTCAATAGATGCTGATAAGAATCACCTGGTGACTGGTGGAGGTGATTGTGCAGTTATACTGCATCCACTAACAACTAATAATGGCATTGTTCACAACATTTTAAACTTTGGAACTCAGAGAAAAGTTATGTTCACTGCGAAAAGAAATTTAATAATGTTGGATGATAAAAATTTGTTGCACCATAACACAAACGAGAAATCATCTAAGGTTTATGAATTAAGTCACGAGTCCACATACAAAATGCTGAGTTTATCATCATGCAAGCAAATTATTGCCGTTGCCGATATGAAAGGAAAACTTGATGTCTTTGTTGAAAACTGTAAGAATGGACACTTAACAAAAATAATTGACACAAAATTGCAATTAGGAAAGATATTATCAATGCAGTGGGCTGGCAACAGACATGTAATTATTTGTTCTGAAAATGGTGCAATAACAGTGTTAGCTGCAAATGGGGAAAATACAGAAGTTTATGCCAATTTCATTTTGCCTCCTTGCAAAGAAAGATGGCTTACGGCCACAGCTTTAAACAATAAAAATGATGTATTTGCTTTAGGAGACAGGTGTGGTAGTATCCATATTTATGTTAAAGGAAAATGTAATCCAATCAAAAGCTTTAAGAAAGTACACGGAAGATATGGTCCAACATCAATTAAATTTAACAGCAATGAACTTGTGTCAACCGGAAGAGATGGGACAATCAAATATTTCAAAATCGATATTCACAACGAAATTATATTTATGAGGAGTAGGGTCTTAGAGTTTCAGTGGGTTGAGGAATTCTTGAATACCGATGAAACATTGATATGTGGTTTTCAAGAAAGACTTTTTGTGATTTATGACCTGACAGACAACTCCAAAGTACTTGAAGTGACATGTGGAGGAGGTCACAGGTCGTGGGACGCAGTGCGTTTCATAGAGAAAGTCAATGACGACTTCGAACAATTTGTTAATTTCATTTATTTGAAAAATTCTGAAATACATCTATTGAAGTTTCAACTGAGTAAAATTGTGGCAAAGAATTTAGTGAACGGATCACATTCTAACAACATAAACTGCTTAAAAGTCTTAGTTAGTGATAGATCGAAAACAATTTACGTATCAGGGGGAGAAGACACAACAGTAAGAATATCGACCACAGACAATCAACTCAATTTTAAAGACGTCTTAACATTTAAACAGCTGTCAAACGTCAGAACCCTTAAATTGCAACCTGTAGATGACAACAATAGTGTAATTTTTACAGCTGGAGGCCGTGCACAAATATGTATAAAAAAATGTACTATAAAAGAAAATGAAATTAAGACCCAAGATTTAATTGAGTACATGATAAAGGGTACAGATAAGGAGAGGAAAGGTAATCAGACGTGGCGGAATTGCACAATAGATTGCGATCCAGAGACAAGGATCATGGATATTGCAGTTTTGAAGAATACTGATTACTTCATTTTCACCGGATGTTCCGACGCTTGTCTGCGAGTTTTGAAATATGAATTACTAATCAATAAAATAACATTAATCAATACAATTAGTCACCATGACACATGTATTTTAAAAACCCTTTGTTTCCGGCTGTTGGACAGAGACTTCTTGGTGACAGCCACAACAAGAGGTCTGTTCTCGATATGGCTAGCTTCTGACCTGCTGAAAGAGAACCCTGCGCCGTTTTTCTCAATAAAAACGAACAAATCCGGAATAAATAGTATAAGTTGTTATAATATATCAAAGAACGAAGTTTTACTAGCCACGGCAGGTGATGATAATACCATCTACATGACATCAATAGAGATAAACATCCAAGATAACTTGCCCTCGGCGGTCGTCAAATATTGTTGGAGCTCTGATCAGTTTCACAGCTCCCAGATAACTGGTTTATGGTTGGGCAATGGCTTGCTACTCAGTACATCGGTCGACCAGAGGGTAACTATGTACAGCTGGACTGTTGATGACGGTTTACACTGCTGCTGTCTACGGCAAGGTGTTAGCGACGTGGCTGACATACAAGGGATGGAAATGCTGGAATGTACGAGTGATCACATAAAGCTGTGTGTCTTCGGCAAAGGAATGGAAGTCCTTCAGCTGCAACATACAAACACATAA
Protein
MSLLIRTDVTAVKIFKNYVLAGIGSTINVFDKASARLITKFQCLKSQKIYGFVVSNCGNKILVFGGKQFTVLSFTHSHYEETFKRNFEPIICDDWLHSGVWLKEDIVILLTAHNVTQKWNLSTQSFISQEAGKENSILYSGLIVMLQDAIMVLAGTVFSDVIVHSNTVKEPLHYLRGHKGVIFSISCDTDKGLIVTTSDDRSVRIWSTNIITANAHMLQYWKDHEITCKYELYGHSARVMRNCITNNIVISVGEDSAICFWNLNGELIKKNNSHQYSGIWSIDADKNHLVTGGGDCAVILHPLTTNNGIVHNILNFGTQRKVMFTAKRNLIMLDDKNLLHHNTNEKSSKVYELSHESTYKMLSLSSCKQIIAVADMKGKLDVFVENCKNGHLTKIIDTKLQLGKILSMQWAGNRHVIICSENGAITVLAANGENTEVYANFILPPCKERWLTATALNNKNDVFALGDRCGSIHIYVKGKCNPIKSFKKVHGRYGPTSIKFNSNELVSTGRDGTIKYFKIDIHNEIIFMRSRVLEFQWVEEFLNTDETLICGFQERLFVIYDLTDNSKVLEVTCGGGHRSWDAVRFIEKVNDDFEQFVNFIYLKNSEIHLLKFQLSKIVAKNLVNGSHSNNINCLKVLVSDRSKTIYVSGGEDTTVRISTTDNQLNFKDVLTFKQLSNVRTLKLQPVDDNNSVIFTAGGRAQICIKKCTIKENEIKTQDLIEYMIKGTDKERKGNQTWRNCTIDCDPETRIMDIAVLKNTDYFIFTGCSDACLRVLKYELLINKITLINTISHHDTCILKTLCFRLLDRDFLVTATTRGLFSIWLASDLLKENPAPFFSIKTNKSGINSISCYNISKNEVLLATAGDDNTIYMTSIEINIQDNLPSAVVKYCWSSDQFHSSQITGLWLGNGLLLSTSVDQRVTMYSWTVDDGLHCCCLRQGVSDVADIQGMEMLECTSDHIKLCVFGKGMEVLQLQHTNT
Summary
Uniprot
H9JP16
A0A2A4JJV2
A0A2H1WUE9
A0A3S2NMN6
A0A212FHS8
A0A0L7LCI2
+ More
A0A2J7PV43 A0A0V0G9P1 A0A023EY21 A0A0J7KWU4 A0A3L8D9J6 A0A2J7PV31 A0A026X2L4 A0A0M8ZX17 A0A088ARN9 A0A2A3E2D0 F4W7E9 A0A151X8M7 A0A154PBE6 A0A232F1A7 E2BQG6 A0A158NPU5 A0A151JSQ9 A0A0L7RDE1 A0A195AU16 E2AVE6 A0A1Y1M087 A0A1W4WWL1 A0A195DSU6 A0A1Y1LY20 A0A151I6V2 A0A1B6D4L0 E0VWF9 A0A067R4E2 A0A146L868 A0A0A9WDW8 A0A0P6E8U5 A0A0P5BU59 A0A1B6HZJ5 A0A1W4X613 E9GLN9 A0A0N8B047 A0A0P5T1U0 A0A0P5FF63 A0A1Y1M098 A0A0P6BKK0 A0A1Y1LXX7 A0A1Y1LY14 A0A2P8Z9N4 A0A165AG08 V3ZI09 A0A2R5LAT4 A0A1S3IYL4 A0A2L2YF30 K1QY02 H2ZTN8 A0A210PPC9 A0A0L8GQE7 A0A1L8GPR7 A0A2R7W0R2 A9C3T6 A0A2R8QLM0 G1KM73 V8NU63 A0A3N0YSI2 A0A0A1WYF2 A0A034WNQ5 F7CZ88 A0A182W3K2 A0A3B3ZKE2 A0A3B3SVH8 A0A3B3ZKD3 A0A131XUE4 A0A147BNN8 A0A0K8VR24 A0A0K8VMX7 A0A0K8W1Y8 A0A182Y8N3 W8BXL6 M7B3B0 A0A182XIE5 Q7Q9X9 A0A2C9KQE6 A0A1B0G1R5 A0A182JUY7 A0A034WQF0 A0A182PHX8 A0A182HYT2 F6ZLR0 A0A0P5HNR7 A0A034WK67 A0A1B0ABY4 A0A1B0ARY4 A0A1A9Y1E5 H2THW1 A0A1I8PBN5 T1JG63 Q17HU0
A0A2J7PV43 A0A0V0G9P1 A0A023EY21 A0A0J7KWU4 A0A3L8D9J6 A0A2J7PV31 A0A026X2L4 A0A0M8ZX17 A0A088ARN9 A0A2A3E2D0 F4W7E9 A0A151X8M7 A0A154PBE6 A0A232F1A7 E2BQG6 A0A158NPU5 A0A151JSQ9 A0A0L7RDE1 A0A195AU16 E2AVE6 A0A1Y1M087 A0A1W4WWL1 A0A195DSU6 A0A1Y1LY20 A0A151I6V2 A0A1B6D4L0 E0VWF9 A0A067R4E2 A0A146L868 A0A0A9WDW8 A0A0P6E8U5 A0A0P5BU59 A0A1B6HZJ5 A0A1W4X613 E9GLN9 A0A0N8B047 A0A0P5T1U0 A0A0P5FF63 A0A1Y1M098 A0A0P6BKK0 A0A1Y1LXX7 A0A1Y1LY14 A0A2P8Z9N4 A0A165AG08 V3ZI09 A0A2R5LAT4 A0A1S3IYL4 A0A2L2YF30 K1QY02 H2ZTN8 A0A210PPC9 A0A0L8GQE7 A0A1L8GPR7 A0A2R7W0R2 A9C3T6 A0A2R8QLM0 G1KM73 V8NU63 A0A3N0YSI2 A0A0A1WYF2 A0A034WNQ5 F7CZ88 A0A182W3K2 A0A3B3ZKE2 A0A3B3SVH8 A0A3B3ZKD3 A0A131XUE4 A0A147BNN8 A0A0K8VR24 A0A0K8VMX7 A0A0K8W1Y8 A0A182Y8N3 W8BXL6 M7B3B0 A0A182XIE5 Q7Q9X9 A0A2C9KQE6 A0A1B0G1R5 A0A182JUY7 A0A034WQF0 A0A182PHX8 A0A182HYT2 F6ZLR0 A0A0P5HNR7 A0A034WK67 A0A1B0ABY4 A0A1B0ARY4 A0A1A9Y1E5 H2THW1 A0A1I8PBN5 T1JG63 Q17HU0
Pubmed
19121390
22118469
26227816
25474469
30249741
24508170
+ More
21719571 28648823 20798317 21347285 28004739 20566863 24845553 26823975 25401762 21292972 29403074 23254933 26383154 26561354 22992520 9215903 28812685 27762356 23594743 24297900 25830018 25348373 20431018 25463417 29240929 29652888 25244985 24495485 23624526 12364791 14747013 17210077 15562597 18464734 21551351 17510324
21719571 28648823 20798317 21347285 28004739 20566863 24845553 26823975 25401762 21292972 29403074 23254933 26383154 26561354 22992520 9215903 28812685 27762356 23594743 24297900 25830018 25348373 20431018 25463417 29240929 29652888 25244985 24495485 23624526 12364791 14747013 17210077 15562597 18464734 21551351 17510324
EMBL
BABH01009639
NWSH01001330
PCG71672.1
ODYU01011140
SOQ56695.1
RSAL01000241
+ More
RVE43646.1 AGBW02008466 OWR53291.1 JTDY01001710 KOB73105.1 NEVH01020989 PNF20194.1 GECL01001636 JAP04488.1 GBBI01004729 JAC13983.1 LBMM01002308 KMQ95002.1 QOIP01000011 RLU17006.1 PNF20195.1 KK107021 EZA62333.1 KQ435814 KOX72667.1 KZ288427 PBC25855.1 GL887844 EGI69822.1 KQ982409 KYQ56723.1 KQ434856 KZC08724.1 NNAY01001286 OXU24481.1 GL449769 EFN82024.1 ADTU01022755 KQ982014 KYN30635.1 KQ414614 KOC68853.1 KQ976745 KYM75474.1 GL443090 EFN62580.1 GEZM01044209 JAV78398.1 KQ980487 KYN15921.1 GEZM01044204 JAV78409.1 KQ978456 KYM93895.1 GEDC01016712 JAS20586.1 DS235819 EEB17710.1 KK852759 KDR17043.1 GDHC01014690 JAQ03939.1 GBHO01038896 GBHO01038895 JAG04708.1 JAG04709.1 GDIQ01067735 JAN27002.1 GDIP01180565 JAJ42837.1 GECU01027617 JAS80089.1 GL732551 EFX79538.1 GDIQ01225883 JAK25842.1 GDIP01138917 JAL64797.1 GDIQ01258866 JAJ92858.1 GEZM01044205 JAV78408.1 GDIP01021009 JAM82706.1 GEZM01044203 JAV78412.1 GEZM01044208 JAV78399.1 PYGN01000136 PSN53207.1 LRGB01000626 KZS17594.1 KB204089 ESO81945.1 GGLE01002473 MBY06599.1 IAAA01024919 IAAA01024921 IAAA01024922 LAA06752.1 JH816882 EKC33810.1 AFYH01223649 AFYH01223650 NEDP02005569 OWF38296.1 KQ420924 KOF78845.1 CM004472 OCT85858.1 KK854226 PTY13327.1 CR847503 AZIM01001838 ETE65605.1 RJVU01027559 ROL49169.1 GBXI01010188 JAD04104.1 GAKP01002970 JAC55982.1 AAMC01018812 GEFM01005844 JAP69952.1 GEGO01003015 JAR92389.1 GDHF01010985 JAI41329.1 GDHF01012087 JAI40227.1 GDHF01007464 JAI44850.1 GAMC01002513 JAC04043.1 KB577555 EMP26598.1 AAAB01008898 EAA09303.5 CCAG010007356 GAKP01002969 JAC55983.1 APCN01005404 GDIQ01224676 JAK27049.1 GAKP01002971 JAC55981.1 JXJN01002620 JH432192 CH477245 EAT46252.1
RVE43646.1 AGBW02008466 OWR53291.1 JTDY01001710 KOB73105.1 NEVH01020989 PNF20194.1 GECL01001636 JAP04488.1 GBBI01004729 JAC13983.1 LBMM01002308 KMQ95002.1 QOIP01000011 RLU17006.1 PNF20195.1 KK107021 EZA62333.1 KQ435814 KOX72667.1 KZ288427 PBC25855.1 GL887844 EGI69822.1 KQ982409 KYQ56723.1 KQ434856 KZC08724.1 NNAY01001286 OXU24481.1 GL449769 EFN82024.1 ADTU01022755 KQ982014 KYN30635.1 KQ414614 KOC68853.1 KQ976745 KYM75474.1 GL443090 EFN62580.1 GEZM01044209 JAV78398.1 KQ980487 KYN15921.1 GEZM01044204 JAV78409.1 KQ978456 KYM93895.1 GEDC01016712 JAS20586.1 DS235819 EEB17710.1 KK852759 KDR17043.1 GDHC01014690 JAQ03939.1 GBHO01038896 GBHO01038895 JAG04708.1 JAG04709.1 GDIQ01067735 JAN27002.1 GDIP01180565 JAJ42837.1 GECU01027617 JAS80089.1 GL732551 EFX79538.1 GDIQ01225883 JAK25842.1 GDIP01138917 JAL64797.1 GDIQ01258866 JAJ92858.1 GEZM01044205 JAV78408.1 GDIP01021009 JAM82706.1 GEZM01044203 JAV78412.1 GEZM01044208 JAV78399.1 PYGN01000136 PSN53207.1 LRGB01000626 KZS17594.1 KB204089 ESO81945.1 GGLE01002473 MBY06599.1 IAAA01024919 IAAA01024921 IAAA01024922 LAA06752.1 JH816882 EKC33810.1 AFYH01223649 AFYH01223650 NEDP02005569 OWF38296.1 KQ420924 KOF78845.1 CM004472 OCT85858.1 KK854226 PTY13327.1 CR847503 AZIM01001838 ETE65605.1 RJVU01027559 ROL49169.1 GBXI01010188 JAD04104.1 GAKP01002970 JAC55982.1 AAMC01018812 GEFM01005844 JAP69952.1 GEGO01003015 JAR92389.1 GDHF01010985 JAI41329.1 GDHF01012087 JAI40227.1 GDHF01007464 JAI44850.1 GAMC01002513 JAC04043.1 KB577555 EMP26598.1 AAAB01008898 EAA09303.5 CCAG010007356 GAKP01002969 JAC55983.1 APCN01005404 GDIQ01224676 JAK27049.1 GAKP01002971 JAC55981.1 JXJN01002620 JH432192 CH477245 EAT46252.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000037510
UP000235965
+ More
UP000036403 UP000279307 UP000053097 UP000053105 UP000005203 UP000242457 UP000007755 UP000075809 UP000076502 UP000215335 UP000008237 UP000005205 UP000078541 UP000053825 UP000078540 UP000000311 UP000192223 UP000078492 UP000078542 UP000009046 UP000027135 UP000000305 UP000245037 UP000076858 UP000030746 UP000085678 UP000005408 UP000008672 UP000242188 UP000053454 UP000186698 UP000000437 UP000001646 UP000008143 UP000075920 UP000261520 UP000261540 UP000076408 UP000031443 UP000076407 UP000007062 UP000076420 UP000092444 UP000075881 UP000075885 UP000075840 UP000002279 UP000092445 UP000092460 UP000092443 UP000005226 UP000095300 UP000008820
UP000036403 UP000279307 UP000053097 UP000053105 UP000005203 UP000242457 UP000007755 UP000075809 UP000076502 UP000215335 UP000008237 UP000005205 UP000078541 UP000053825 UP000078540 UP000000311 UP000192223 UP000078492 UP000078542 UP000009046 UP000027135 UP000000305 UP000245037 UP000076858 UP000030746 UP000085678 UP000005408 UP000008672 UP000242188 UP000053454 UP000186698 UP000000437 UP000001646 UP000008143 UP000075920 UP000261520 UP000261540 UP000076408 UP000031443 UP000076407 UP000007062 UP000076420 UP000092444 UP000075881 UP000075885 UP000075840 UP000002279 UP000092445 UP000092460 UP000092443 UP000005226 UP000095300 UP000008820
Interpro
IPR011047
Quinoprotein_ADH-like_supfam
+ More
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001017 DH_E1
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR034616 BCKDH_E1-a
IPR029061 THDP-binding
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR011044 Quino_amine_DH_bsu
IPR011041 Quinoprot_gluc/sorb_DH
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001017 DH_E1
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR034616 BCKDH_E1-a
IPR029061 THDP-binding
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR011044 Quino_amine_DH_bsu
IPR011041 Quinoprot_gluc/sorb_DH
SUPFAM
Gene 3D
ProteinModelPortal
H9JP16
A0A2A4JJV2
A0A2H1WUE9
A0A3S2NMN6
A0A212FHS8
A0A0L7LCI2
+ More
A0A2J7PV43 A0A0V0G9P1 A0A023EY21 A0A0J7KWU4 A0A3L8D9J6 A0A2J7PV31 A0A026X2L4 A0A0M8ZX17 A0A088ARN9 A0A2A3E2D0 F4W7E9 A0A151X8M7 A0A154PBE6 A0A232F1A7 E2BQG6 A0A158NPU5 A0A151JSQ9 A0A0L7RDE1 A0A195AU16 E2AVE6 A0A1Y1M087 A0A1W4WWL1 A0A195DSU6 A0A1Y1LY20 A0A151I6V2 A0A1B6D4L0 E0VWF9 A0A067R4E2 A0A146L868 A0A0A9WDW8 A0A0P6E8U5 A0A0P5BU59 A0A1B6HZJ5 A0A1W4X613 E9GLN9 A0A0N8B047 A0A0P5T1U0 A0A0P5FF63 A0A1Y1M098 A0A0P6BKK0 A0A1Y1LXX7 A0A1Y1LY14 A0A2P8Z9N4 A0A165AG08 V3ZI09 A0A2R5LAT4 A0A1S3IYL4 A0A2L2YF30 K1QY02 H2ZTN8 A0A210PPC9 A0A0L8GQE7 A0A1L8GPR7 A0A2R7W0R2 A9C3T6 A0A2R8QLM0 G1KM73 V8NU63 A0A3N0YSI2 A0A0A1WYF2 A0A034WNQ5 F7CZ88 A0A182W3K2 A0A3B3ZKE2 A0A3B3SVH8 A0A3B3ZKD3 A0A131XUE4 A0A147BNN8 A0A0K8VR24 A0A0K8VMX7 A0A0K8W1Y8 A0A182Y8N3 W8BXL6 M7B3B0 A0A182XIE5 Q7Q9X9 A0A2C9KQE6 A0A1B0G1R5 A0A182JUY7 A0A034WQF0 A0A182PHX8 A0A182HYT2 F6ZLR0 A0A0P5HNR7 A0A034WK67 A0A1B0ABY4 A0A1B0ARY4 A0A1A9Y1E5 H2THW1 A0A1I8PBN5 T1JG63 Q17HU0
A0A2J7PV43 A0A0V0G9P1 A0A023EY21 A0A0J7KWU4 A0A3L8D9J6 A0A2J7PV31 A0A026X2L4 A0A0M8ZX17 A0A088ARN9 A0A2A3E2D0 F4W7E9 A0A151X8M7 A0A154PBE6 A0A232F1A7 E2BQG6 A0A158NPU5 A0A151JSQ9 A0A0L7RDE1 A0A195AU16 E2AVE6 A0A1Y1M087 A0A1W4WWL1 A0A195DSU6 A0A1Y1LY20 A0A151I6V2 A0A1B6D4L0 E0VWF9 A0A067R4E2 A0A146L868 A0A0A9WDW8 A0A0P6E8U5 A0A0P5BU59 A0A1B6HZJ5 A0A1W4X613 E9GLN9 A0A0N8B047 A0A0P5T1U0 A0A0P5FF63 A0A1Y1M098 A0A0P6BKK0 A0A1Y1LXX7 A0A1Y1LY14 A0A2P8Z9N4 A0A165AG08 V3ZI09 A0A2R5LAT4 A0A1S3IYL4 A0A2L2YF30 K1QY02 H2ZTN8 A0A210PPC9 A0A0L8GQE7 A0A1L8GPR7 A0A2R7W0R2 A9C3T6 A0A2R8QLM0 G1KM73 V8NU63 A0A3N0YSI2 A0A0A1WYF2 A0A034WNQ5 F7CZ88 A0A182W3K2 A0A3B3ZKE2 A0A3B3SVH8 A0A3B3ZKD3 A0A131XUE4 A0A147BNN8 A0A0K8VR24 A0A0K8VMX7 A0A0K8W1Y8 A0A182Y8N3 W8BXL6 M7B3B0 A0A182XIE5 Q7Q9X9 A0A2C9KQE6 A0A1B0G1R5 A0A182JUY7 A0A034WQF0 A0A182PHX8 A0A182HYT2 F6ZLR0 A0A0P5HNR7 A0A034WK67 A0A1B0ABY4 A0A1B0ARY4 A0A1A9Y1E5 H2THW1 A0A1I8PBN5 T1JG63 Q17HU0
PDB
6G6N
E-value=0.000309981,
Score=108
Ontologies
KEGG
GO
PANTHER
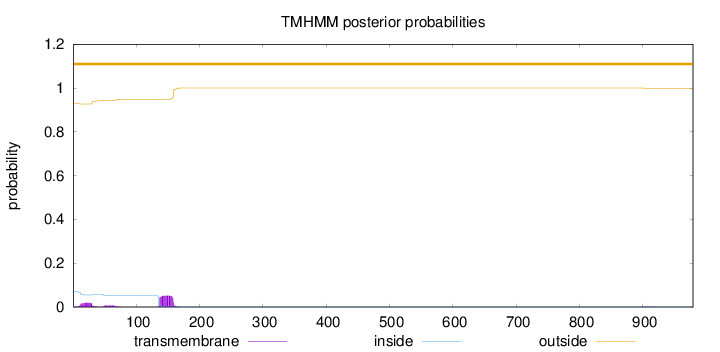
Topology
Length:
979
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.68409000000001
Exp number, first 60 AAs:
0.45365
Total prob of N-in:
0.07055
outside
1 - 979
Population Genetic Test Statistics
Pi
238.572304
Theta
202.46659
Tajima's D
0.610361
CLR
0.334965
CSRT
0.544872756362182
Interpretation
Uncertain
