Gene
KWMTBOMO13985 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011271
Annotation
PREDICTED:_regucalcin-like_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.323
Sequence
CDS
ATGGCGCCGCAGTTACAAGCTGTGACGGAGCCGGTGTGGCTCGGCGAGGGCCCACACTGGGACCACAACGAGCAGGCCCTATACTTTGTCAGCATCTTCGACGAGACCATACATAAGTACGTGCCAGAAACGGGGAAACATACGAGGTCTAAACTAGACGGGAAGCCAACCTTCATTATACCCATAGAAGGGAAGAAACACCATTTCGTGGTGGGTCTGGACAGGCTGGTCGTGGAGATACAATGGACAGGAGAAGACCAGACGGCCAGGCTCGTCAGGACCGTCGCGGAGGTGGACCAGGACAACCCCAACAATAGATTCAACGACGCTAAAGCTGACCCCAGGGGCAGGCTGTTTGCCGGAACGATGGGTCACGAATACGAACCGGGAAAGTTCGACCTGAAGAAGGGGTCTCTGTACCGCATCGACCCGGACGGCTCCGTGCACCGCCTCGAGTCCAACATCGACATCTCCAACGGCCTCTGCTGGGACCTGCAGCGGAGCGCCTTCTACTTCGCGGACTCCTTCGAGTACACCATCAGGAGATACGACTACGACGTGGAGACCGGGAGCATCGCGAACGCAAAGACCGTGTTTAAGTACAGCGACCACGGACTGGAAGGTATCGTGGACGGCATGACCATAGACACGGACGGTAACCTCTGGGTGGCCAACTTTGATGGACGACAGGTGTTAAAGATCGATCCTCGTGGCGGCAAACTTCTGCAGAAGATCCCGATACCGGCGCTCCAAGTCACGTCAGTGACATTCGGGGGTCCGGACCTGGACCAGTTGTACGTGACCTCCGCCTCTATGAACCGCGGCGAAGAACAACTCCCGCCCTGCGGCTCTACCTTTAGACTGGACGGACTCGAGGTCAAAGGACATCCTAATGTGAACGTGCGTCTTCAGTGA
Protein
MAPQLQAVTEPVWLGEGPHWDHNEQALYFVSIFDETIHKYVPETGKHTRSKLDGKPTFIIPIEGKKHHFVVGLDRLVVEIQWTGEDQTARLVRTVAEVDQDNPNNRFNDAKADPRGRLFAGTMGHEYEPGKFDLKKGSLYRIDPDGSVHRLESNIDISNGLCWDLQRSAFYFADSFEYTIRRYDYDVETGSIANAKTVFKYSDHGLEGIVDGMTIDTDGNLWVANFDGRQVLKIDPRGGKLLQKIPIPALQVTSVTFGGPDLDQLYVTSASMNRGEEQLPPCGSTFRLDGLEVKGHPNVNVRLQ
Summary
Uniprot
H9JP17
A0A2A4JI81
I4DLB5
A0A0N1I833
A0A212FHS6
A0A194QNP8
+ More
A0A3S2LTM3 A0A2Z5U7W2 I4DP01 A0A0L7L6D7 H9JP83 A0A0N0PBN4 I4DLC0 A0A2A4JIX8 A0A0L7L6J1 A0A2H1WUP7 A0A194QJM9 A0A194RFD5 A0A212FHT2 A0A0N1IES4 A0A194QNQ3 A0A194QHS8 A0A194QI87 E0XEM8 A0A194QHX1 A0A0N1IP04 A0A194QJM0 A0A2J7PV34 A0A1Y1KUS1 J3JZF0 D6WJC9 A0A139WHG8 A0A2P8Z9M7 U4TVX1 A0A2J7PV36 M9NHL6 A0A1B6LND9 A0A1B6DWB3 W8ANX0 A0A0Q9X2K6 A0A1B6G0U5 A0A1B6GG13 B4L5N8 D6WJC6 A0A0Q9XH70 V5I8P9 M4QVG7 M9NK07 A0A1B0DL94 B4MG80 A0A1D2NDD8 M4QII6 M4QKU5 A0A0Q9VYG4 A0A1L8DLY9 A0A084VI31 A0A0Q9X3C4 A0A1L8DM86 A0A336KBK1 A0A336KH96 M4QIK2 A0A1B6JPW2 J9LYB7 M4QID0 A0A1B6J451 B3MXT5 A0A0P8ZSH9 A0A0P8XYJ7 B3LWR9 A0A0P9AQK9 Q9VYR1 Q76NR6 A0A0Q5TC32 N6TMF5 A0A182IEG6 C6TP78 A0A0R1EAS6 A0A0N1IGA3 B4PYN1 A0A1B6D0K5 Q9NDP1 D6WJC8 A0A182L2J6 A0A182X1I5 A0A0P8XZ51 B3NXM6 Q16IH7 J9JXU7 A0A182IYV2 A0A1W4VZ14 B4R3K9 E7BDF0 A0A1B6EIG4 E7BDE9 A0A1A8CI06 A0A1A8IYK7 A0A1A8AP47 A0A1S4GW88 A0A1W4XLV1 B4ILA7
A0A3S2LTM3 A0A2Z5U7W2 I4DP01 A0A0L7L6D7 H9JP83 A0A0N0PBN4 I4DLC0 A0A2A4JIX8 A0A0L7L6J1 A0A2H1WUP7 A0A194QJM9 A0A194RFD5 A0A212FHT2 A0A0N1IES4 A0A194QNQ3 A0A194QHS8 A0A194QI87 E0XEM8 A0A194QHX1 A0A0N1IP04 A0A194QJM0 A0A2J7PV34 A0A1Y1KUS1 J3JZF0 D6WJC9 A0A139WHG8 A0A2P8Z9M7 U4TVX1 A0A2J7PV36 M9NHL6 A0A1B6LND9 A0A1B6DWB3 W8ANX0 A0A0Q9X2K6 A0A1B6G0U5 A0A1B6GG13 B4L5N8 D6WJC6 A0A0Q9XH70 V5I8P9 M4QVG7 M9NK07 A0A1B0DL94 B4MG80 A0A1D2NDD8 M4QII6 M4QKU5 A0A0Q9VYG4 A0A1L8DLY9 A0A084VI31 A0A0Q9X3C4 A0A1L8DM86 A0A336KBK1 A0A336KH96 M4QIK2 A0A1B6JPW2 J9LYB7 M4QID0 A0A1B6J451 B3MXT5 A0A0P8ZSH9 A0A0P8XYJ7 B3LWR9 A0A0P9AQK9 Q9VYR1 Q76NR6 A0A0Q5TC32 N6TMF5 A0A182IEG6 C6TP78 A0A0R1EAS6 A0A0N1IGA3 B4PYN1 A0A1B6D0K5 Q9NDP1 D6WJC8 A0A182L2J6 A0A182X1I5 A0A0P8XZ51 B3NXM6 Q16IH7 J9JXU7 A0A182IYV2 A0A1W4VZ14 B4R3K9 E7BDF0 A0A1B6EIG4 E7BDE9 A0A1A8CI06 A0A1A8IYK7 A0A1A8AP47 A0A1S4GW88 A0A1W4XLV1 B4ILA7
Pubmed
19121390
22651552
26354079
22118469
26227816
28004739
+ More
22516182 18362917 19820115 29403074 23537049 24495485 17994087 18057021 23493635 27289101 24438588 10788801 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20966253 17510324 21339509 12364791
22516182 18362917 19820115 29403074 23537049 24495485 17994087 18057021 23493635 27289101 24438588 10788801 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20966253 17510324 21339509 12364791
EMBL
BABH01009637
NWSH01001330
PCG71671.1
AK402083
BAM18705.1
KQ460880
+ More
KPJ11522.1 AGBW02008466 OWR53292.1 KQ458761 KPJ05106.1 RSAL01000241 RVE43645.1 AP017497 BBB06781.1 AK403222 BAM19641.1 JTDY01002624 KOB71073.1 KPJ11518.1 AK402088 BAM18710.1 PCG71668.1 KOB71072.1 ODYU01011140 SOQ56696.1 KPJ05120.1 KQ460297 KPJ16169.1 OWR53293.1 KPJ11521.1 KPJ05111.1 KPJ05108.1 KPJ05109.1 GU013474 ADK55065.1 KPJ05107.1 KPJ11519.1 KPJ05110.1 NEVH01020989 PNF20197.1 GEZM01075834 GEZM01075833 JAV63930.1 BT128631 AEE63588.1 KQ971343 EFA03133.1 KYB27438.1 PYGN01000136 PSN53206.1 KB631643 ERL84922.1 PNF20199.1 JN036578 AFJ24877.1 GEBQ01028432 GEBQ01014707 GEBQ01009166 GEBQ01008064 JAT11545.1 JAT25270.1 JAT30811.1 JAT31913.1 GEDC01007379 JAS29919.1 GAMC01020362 GAMC01020361 JAB86193.1 CH964232 KRF99329.1 GECZ01013723 JAS56046.1 GECZ01008401 JAS61368.1 CH933811 EDW06497.2 KRG07000.1 EFA03136.1 KRG07001.1 GALX01004305 JAB64161.1 KC689140 AGH29386.1 JN036579 AFJ24878.1 AJVK01016346 AJVK01016347 CH940672 EDW57403.2 LJIJ01000079 ODN03278.1 KC689138 KC689139 KC689142 KC689143 KC689145 KC689146 KC689147 KC689148 KC689149 KC689150 KC689151 KC689152 KC689154 KC689155 KC689156 AGH29384.1 AGH29385.1 AGH29388.1 AGH29389.1 AGH29391.1 AGH29392.1 AGH29393.1 AGH29394.1 AGH29395.1 AGH29396.1 AGH29397.1 AGH29398.1 AGH29400.1 AGH29401.1 AGH29402.1 KC689141 AGH29387.1 KRF77757.1 GFDF01006606 JAV07478.1 ATLV01013269 KE524849 KFB37625.1 KRF99328.1 GFDF01006607 JAV07477.1 UFQS01000275 UFQT01000275 SSX02284.1 SSX22660.1 SSX02283.1 SSX22659.1 KC689153 AGH29399.1 GECU01006486 JAT01221.1 ABLF02034011 KC689144 AGH29390.1 GECU01013752 JAS93954.1 CH902630 EDV38550.1 KPU77449.1 KPU77448.1 CH902617 KPU79867.1 EDV42707.2 KPU79865.1 AE014298 AY118643 AB036904 AAF48128.1 AAF48129.1 AAF48130.1 AAM50012.1 BAA99283.1 AAN09306.2 CH954180 KQS30449.1 APGK01018473 APGK01018474 KB740076 ENN81654.1 APCN01005203 BT099564 ACU33956.1 CM000162 KRK05853.1 KRK05854.1 KPJ11520.1 EDX00967.1 GEDC01018111 JAS19187.1 AB036903 BAA99282.1 EFA03134.2 KPU79866.1 EDV47327.2 CH478076 EAT34066.1 ABLF02026048 CM000366 EDX17711.1 FR774042 CBY89805.1 GECZ01032052 JAS37717.1 FR774041 CBY89804.1 HADZ01015408 SBP79349.1 HAED01015989 SBR02434.1 HADY01018324 SBP56809.1 AAAB01008964 CH480868 EDW53753.1
KPJ11522.1 AGBW02008466 OWR53292.1 KQ458761 KPJ05106.1 RSAL01000241 RVE43645.1 AP017497 BBB06781.1 AK403222 BAM19641.1 JTDY01002624 KOB71073.1 KPJ11518.1 AK402088 BAM18710.1 PCG71668.1 KOB71072.1 ODYU01011140 SOQ56696.1 KPJ05120.1 KQ460297 KPJ16169.1 OWR53293.1 KPJ11521.1 KPJ05111.1 KPJ05108.1 KPJ05109.1 GU013474 ADK55065.1 KPJ05107.1 KPJ11519.1 KPJ05110.1 NEVH01020989 PNF20197.1 GEZM01075834 GEZM01075833 JAV63930.1 BT128631 AEE63588.1 KQ971343 EFA03133.1 KYB27438.1 PYGN01000136 PSN53206.1 KB631643 ERL84922.1 PNF20199.1 JN036578 AFJ24877.1 GEBQ01028432 GEBQ01014707 GEBQ01009166 GEBQ01008064 JAT11545.1 JAT25270.1 JAT30811.1 JAT31913.1 GEDC01007379 JAS29919.1 GAMC01020362 GAMC01020361 JAB86193.1 CH964232 KRF99329.1 GECZ01013723 JAS56046.1 GECZ01008401 JAS61368.1 CH933811 EDW06497.2 KRG07000.1 EFA03136.1 KRG07001.1 GALX01004305 JAB64161.1 KC689140 AGH29386.1 JN036579 AFJ24878.1 AJVK01016346 AJVK01016347 CH940672 EDW57403.2 LJIJ01000079 ODN03278.1 KC689138 KC689139 KC689142 KC689143 KC689145 KC689146 KC689147 KC689148 KC689149 KC689150 KC689151 KC689152 KC689154 KC689155 KC689156 AGH29384.1 AGH29385.1 AGH29388.1 AGH29389.1 AGH29391.1 AGH29392.1 AGH29393.1 AGH29394.1 AGH29395.1 AGH29396.1 AGH29397.1 AGH29398.1 AGH29400.1 AGH29401.1 AGH29402.1 KC689141 AGH29387.1 KRF77757.1 GFDF01006606 JAV07478.1 ATLV01013269 KE524849 KFB37625.1 KRF99328.1 GFDF01006607 JAV07477.1 UFQS01000275 UFQT01000275 SSX02284.1 SSX22660.1 SSX02283.1 SSX22659.1 KC689153 AGH29399.1 GECU01006486 JAT01221.1 ABLF02034011 KC689144 AGH29390.1 GECU01013752 JAS93954.1 CH902630 EDV38550.1 KPU77449.1 KPU77448.1 CH902617 KPU79867.1 EDV42707.2 KPU79865.1 AE014298 AY118643 AB036904 AAF48128.1 AAF48129.1 AAF48130.1 AAM50012.1 BAA99283.1 AAN09306.2 CH954180 KQS30449.1 APGK01018473 APGK01018474 KB740076 ENN81654.1 APCN01005203 BT099564 ACU33956.1 CM000162 KRK05853.1 KRK05854.1 KPJ11520.1 EDX00967.1 GEDC01018111 JAS19187.1 AB036903 BAA99282.1 EFA03134.2 KPU79866.1 EDV47327.2 CH478076 EAT34066.1 ABLF02026048 CM000366 EDX17711.1 FR774042 CBY89805.1 GECZ01032052 JAS37717.1 FR774041 CBY89804.1 HADZ01015408 SBP79349.1 HAED01015989 SBR02434.1 HADY01018324 SBP56809.1 AAAB01008964 CH480868 EDW53753.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000283053
+ More
UP000037510 UP000235965 UP000007266 UP000245037 UP000030742 UP000007798 UP000009192 UP000092462 UP000008792 UP000094527 UP000030765 UP000007819 UP000007801 UP000000803 UP000008711 UP000019118 UP000075840 UP000002282 UP000075882 UP000076407 UP000008820 UP000075880 UP000192221 UP000000304 UP000192223 UP000001292
UP000037510 UP000235965 UP000007266 UP000245037 UP000030742 UP000007798 UP000009192 UP000092462 UP000008792 UP000094527 UP000030765 UP000007819 UP000007801 UP000000803 UP000008711 UP000019118 UP000075840 UP000002282 UP000075882 UP000076407 UP000008820 UP000075880 UP000192221 UP000000304 UP000192223 UP000001292
Interpro
Gene 3D
ProteinModelPortal
H9JP17
A0A2A4JI81
I4DLB5
A0A0N1I833
A0A212FHS6
A0A194QNP8
+ More
A0A3S2LTM3 A0A2Z5U7W2 I4DP01 A0A0L7L6D7 H9JP83 A0A0N0PBN4 I4DLC0 A0A2A4JIX8 A0A0L7L6J1 A0A2H1WUP7 A0A194QJM9 A0A194RFD5 A0A212FHT2 A0A0N1IES4 A0A194QNQ3 A0A194QHS8 A0A194QI87 E0XEM8 A0A194QHX1 A0A0N1IP04 A0A194QJM0 A0A2J7PV34 A0A1Y1KUS1 J3JZF0 D6WJC9 A0A139WHG8 A0A2P8Z9M7 U4TVX1 A0A2J7PV36 M9NHL6 A0A1B6LND9 A0A1B6DWB3 W8ANX0 A0A0Q9X2K6 A0A1B6G0U5 A0A1B6GG13 B4L5N8 D6WJC6 A0A0Q9XH70 V5I8P9 M4QVG7 M9NK07 A0A1B0DL94 B4MG80 A0A1D2NDD8 M4QII6 M4QKU5 A0A0Q9VYG4 A0A1L8DLY9 A0A084VI31 A0A0Q9X3C4 A0A1L8DM86 A0A336KBK1 A0A336KH96 M4QIK2 A0A1B6JPW2 J9LYB7 M4QID0 A0A1B6J451 B3MXT5 A0A0P8ZSH9 A0A0P8XYJ7 B3LWR9 A0A0P9AQK9 Q9VYR1 Q76NR6 A0A0Q5TC32 N6TMF5 A0A182IEG6 C6TP78 A0A0R1EAS6 A0A0N1IGA3 B4PYN1 A0A1B6D0K5 Q9NDP1 D6WJC8 A0A182L2J6 A0A182X1I5 A0A0P8XZ51 B3NXM6 Q16IH7 J9JXU7 A0A182IYV2 A0A1W4VZ14 B4R3K9 E7BDF0 A0A1B6EIG4 E7BDE9 A0A1A8CI06 A0A1A8IYK7 A0A1A8AP47 A0A1S4GW88 A0A1W4XLV1 B4ILA7
A0A3S2LTM3 A0A2Z5U7W2 I4DP01 A0A0L7L6D7 H9JP83 A0A0N0PBN4 I4DLC0 A0A2A4JIX8 A0A0L7L6J1 A0A2H1WUP7 A0A194QJM9 A0A194RFD5 A0A212FHT2 A0A0N1IES4 A0A194QNQ3 A0A194QHS8 A0A194QI87 E0XEM8 A0A194QHX1 A0A0N1IP04 A0A194QJM0 A0A2J7PV34 A0A1Y1KUS1 J3JZF0 D6WJC9 A0A139WHG8 A0A2P8Z9M7 U4TVX1 A0A2J7PV36 M9NHL6 A0A1B6LND9 A0A1B6DWB3 W8ANX0 A0A0Q9X2K6 A0A1B6G0U5 A0A1B6GG13 B4L5N8 D6WJC6 A0A0Q9XH70 V5I8P9 M4QVG7 M9NK07 A0A1B0DL94 B4MG80 A0A1D2NDD8 M4QII6 M4QKU5 A0A0Q9VYG4 A0A1L8DLY9 A0A084VI31 A0A0Q9X3C4 A0A1L8DM86 A0A336KBK1 A0A336KH96 M4QIK2 A0A1B6JPW2 J9LYB7 M4QID0 A0A1B6J451 B3MXT5 A0A0P8ZSH9 A0A0P8XYJ7 B3LWR9 A0A0P9AQK9 Q9VYR1 Q76NR6 A0A0Q5TC32 N6TMF5 A0A182IEG6 C6TP78 A0A0R1EAS6 A0A0N1IGA3 B4PYN1 A0A1B6D0K5 Q9NDP1 D6WJC8 A0A182L2J6 A0A182X1I5 A0A0P8XZ51 B3NXM6 Q16IH7 J9JXU7 A0A182IYV2 A0A1W4VZ14 B4R3K9 E7BDF0 A0A1B6EIG4 E7BDE9 A0A1A8CI06 A0A1A8IYK7 A0A1A8AP47 A0A1S4GW88 A0A1W4XLV1 B4ILA7
PDB
5XFE
E-value=6.21414e-60,
Score=583
Ontologies
PATHWAY
GO
PANTHER
Topology
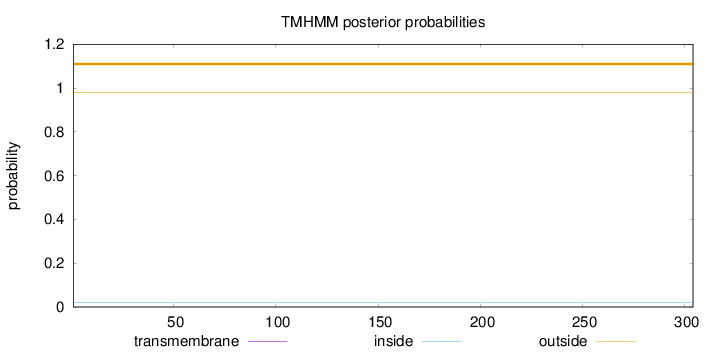
Length:
304
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00044
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02083
outside
1 - 304
Population Genetic Test Statistics
Pi
17.625212
Theta
31.779347
Tajima's D
-1.689037
CLR
0.619528
CSRT
0.0393480325983701
Interpretation
Uncertain
